ABSTRACT
Background: Dietary intake of antioxidants such as vitamins C and E protect against oxidative stress, and may also be associated with altered DNA methylation patterns.
Methods: We meta-analysed epigenome-wide association study (EWAS) results from 11,866 participants across eight population-based cohorts to evaluate the association between self-reported dietary and supplemental intake of vitamins C and E with DNA methylation. EWAS were adjusted for age, sex, BMI, caloric intake, blood cell type proportion, smoking status, alcohol consumption, and technical covariates. Significant results of the meta-analysis were subsequently evaluated in gene set enrichment analysis (GSEA) and expression quantitative trait methylation (eQTM) analysis.
Results: In meta-analysis, methylation at 4,656 CpG sites was significantly associated with vitamin C intake at FDR ≤ 0.05. The most significant CpG sites associated with vitamin C (at FDR ≤ 0.01) were enriched for pathways associated with systems development and cell signalling in GSEA, and were associated with downstream expression of genes enriched in the immune response in eQTM analysis. Furthermore, methylation at 160 CpG sites was significantly associated with vitamin E intake at FDR ≤ 0.05, but GSEA and eQTM analysis of the top most significant CpG sites associated with vitamin E did not identify significant enrichment of any biological pathways investigated.
Conclusions: We identified significant associations of many CpG sites with vitamin C and E intake, and our results suggest that vitamin C intake may be associated with systems development and the immune response.
Introduction
Methylation of DNA cytosine-phosphate-guanine (CpG) sites reflects a reversible epigenetic modification associated with altered regulation of gene expression. DNA methylation is frequently associated with activation or inactivation of genes and biological pathways, and differential methylation of specific CpG loci has been associated with multiple clinical diseases, including cancer [Citation1–3], diabetes mellitus [Citation4,Citation5], and cardiovascular disease [Citation6]. Oxidative stress, characterized by the production of reactive oxygen species (ROS) that can damage DNA, proteins, and lipids, is one mechanism by which perturbations to DNA methylation patterns can contribute to clinical disease [Citation7–10]. Oxidative stress may upregulate the expression of DNA methyltransferases (DNMTs), which catalyse DNA methylation. Minimizing oxidative stress may help to prevent deleterious DNA methylation patterns and clinical disease.
Dietary intake of antioxidants may provide some protection against oxidative stress [Citation11–14], and may also affect DNA methylation patterns. Vitamin C, or ascorbic acid, is a water-soluble antioxidant that prevents oxidative stress through the reduction of ROS, thereby inhibiting peroxidative processes that can damage cells, DNA, and proteins [Citation15]. Vitamin C is found in the cellular cytosol and extracellular fluid, and it coactivates ten-eleven translocation (TET) enzymes that mediate DNA demethylation via conversion of 5-methylcytosine to 5-hydroxymethylcytosine [Citation16–19]. Vitamin C works synergistically with the lipid-soluble vitamin E [Citation20], or α-tocopherol, another antioxidant that interferes with the propagation of lipid radicals, by reducing oxidized vitamin E and thereby allowing it to continue acting as an antioxidant [Citation21–23].
We sought to evaluate epigenomic patterns associated with dietary intake of vitamins C and E in several large cohorts.
Methods
Study design and population
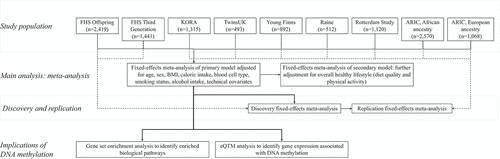
The overall study design is presented in . Cross-sectional data on DNA methylation and vitamin intake from participants enrolled in eight population-based cohorts were used in this study: the Framingham Heart Study (FHS) Offspring cohort (n = 2,419) [Citation24], the FHS Third-Generation cohort (n = 1,441) [Citation25], the UK Adult Twin Registry (TwinsUK; n = 493) [Citation26], the Kooperative Gesundheitsforschung in der Region Augsburg (KORA) study (n = 1,392) [Citation27,Citation28], the Young Finns study (n = 892) [Citation29], Generation 2 of the Australia-based Raine Study (n = 512) [Citation30], the Rotterdam Study (n = 1,120) [Citation31], and the Atherosclerotic Risk in Communities (ARIC) study (n = 2,570 with African ancestry, n = 1,068 with European ancestry) [Citation32]. All cohorts included participants with European ancestry; the ARIC study additionally included participants with African ancestry. Institutional review committees of each cohort approved this study, and all participants provided written informed consent. Data and analytical codes that support our findings are available from the corresponding author upon request. Detailed cohort descriptions for each of these cohorts are presented in the Supplemental Materials.
Clinical and behavioural risk factors
Clinical and behavioural risk factors included as covariates in statistical models were collected according to each cohort’s specific protocols, as detailed in the Supplemental Materials. Body mass index (BMI) was calculated as the ratio of the participant’s measured weight (in kg) to height (in m) squared. Smoking status was obtained based on self-reported data, and participants were categorized as current, former, and never smokers. Self-reported physical activity data were available for the FHS Offspring study, FHS Third-Generation study, YFS, Rotterdam study, and ARIC study and are described in detail in the Supplemental Materials.
Vitamin intake
Dietary and supplementary intake of vitamin C, dietary intake of vitamin E, and overall caloric intake were calculated based on self-reported food frequency questionnaire (FFQ) data or a combination of repeated 24-hour recalls and a FFQ (KORA FF4). The FHS cohorts used the 126-item Harvard-Willett FFQ [Citation33]; the TwinsUK cohort used the 131-item European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk FFQ [Citation34]; the KORA cohort used two 24-hour recalls and the Bavarian Food Consumption Survey [Citation35]; the Young Finns Study cohort used a 131-item FFQ developed by the Finnish National Institute for Health and Welfare [Citation36]; the Raine study used a 212-item FFQ developed by the Commonwealth Scientific and Industrial Research Organisation [Citation37]; the Rotterdam Study used a 389-item Dutch FFQ [Citation38]; and the ARIC cohorts used a 66-item modified Willett FFQ [Citation33,Citation39]. Additional details about the study-specific FFQs are described in the Supplementary Materials.
Continuous vitamin C intake based on dietary and supplemental intake was categorized into low, medium, and high intake based on sex-specific thresholds for adults recommended by the Institute of Medicine of the National Academies (Supplemental Table S1) [Citation40]. Low vitamin C intake was categorized as intake not meeting the recommended dietary allowance; medium vitamin C intake was categorized as intake meeting the recommended dietary allowance up to 300 mg in both men and women; and high vitamin C intake was categorized as any intake above 300 mg in both men and women.
Continuous vitamin E intake based on dietary and supplemental intake was similarly categorized into low, medium, and high intake based on the recommended dietary allowance for vitamin E (Supplemental Table S1) [Citation40]. Low vitamin E intake was categorized as intake not meeting the recommended dietary allowance, medium vitamin E intake was categorized as intake meeting the recommended dietary allowance of 15 mg up to 22.5 mg; and high vitamin E intake was categorized as any intake above 22.5 mg.
Diet quality was assessed in all cohorts included in the secondary model by calculating the Mediterranean diet score [Citation41]. Study-specific FFQs are detailed in the Supplementary Materials.
DNA methylation
DNA methylation measurements were performed independently by each cohort. Fasting whole blood samples were collected from 11,866 participants. Samples for all cohorts except the Young Finns Study cohort were assayed using the Infinium HumanMethylation450 Beadchip (Illumina Inc., San Diego, CA), which covers >450,000 CpG sites. The Young Finns Study cohort samples were assayed using the MethylationEPIC 850K Beadchip (Illumina Inc., San Diego, CA), which covers >850,000 CpG sites. Each cohort normalized methylation beta values; details of the statistical methods for normalization are presented in the Supplemental Materials.
Cohort-specific statistical models
Statistical models evaluated global associations of DNA methylation with vitamin intake by estimating the locus-by-locus association of self-reported vitamin intake with medium and high vitamin intake compared with low intake. All eight cohorts independently tested the association of dietary intake of vitamin C with DNA methylation using linear mixed models. Seven cohorts (the FHS Offspring study, the FHS Third-Generation study, the KORA study, the Young Finns study, the Raine Study, the Rotterdam Study, and the ARIC study) independently tested the association of dietary intake of vitamin E with DNA methylation using linear mixed models.
All statistical models were adjusted for age, sex, BMI, daily caloric intake, blood cell type proportion, smoking status (current/former/never), alcohol consumption, and assay-specific technical covariates. Blood cell type proportions (CD4+ T-cells, CD8+ T-cells, NK cells, monocytes, and eosinophils) were estimated for all participants based on DNA methylation values using the Houseman imputation method [Citation42]. Cohort-specific technical covariates are detailed in the Supplemental Materials. To evaluate whether any significant associations were independent of overall healthy behaviour, a second model further adjusted for physical activity and diet quality. Data from all cohorts were included in both models with the exception of the KORA study cohort, which was not included in the second model due to unavailability of diet quality data at that time.
Epigenome-wide meta-analysis
Inverse variance-weighted fixed-effects meta-analysis was used to combine cohort-specific results. Only CpG sites available in ≥3 cohorts were included in the meta-analysis: for the association between vitamin C intake and DNA methylation, 485,505 CpG sites were included in the meta-analysis, and for the association between vitamin E intake and DNA methylation, 485,433 CpG sites were included in the meta-analysis. The false discovery rate (FDR) was calculated for all models [Citation43], and statistical significance was assessed at a FDR < 0.05.
Discovery and replication
Due to differences in FFQs used to capture dietary and supplemental vitamin intake as well as potential differences in dietary patterns, we divided the total sample across cohorts into separate discovery and replication cohorts to investigate heterogeneity in the meta-analysis results and to assess the robustness of the topmost significant signals in our overall meta-analysis. We evaluated whether associations in the primary model were reproducible in the replication set. The discovery meta-analysis consisted of the 3,833 FHS Offspring and Third-Generation participants since both cohorts used the 126-item Harvard Willet FFQ and the same methods. The replication meta-analysis included 6,655 participants and consisted of the TwinsUK, Young Finns, the Rotterdam Study and ARIC study cohorts. Statistical significance at a FDR ≤ 0.05 was assessed in the replication cohort.
Implications of DNA methylation: gene set enrichment analysis (GSEA) and expression quantitative trait methylation (eQTM) analysis
Genes to which CpG sites associated with vitamin intake were annotated were subsequently used in gene set enrichment analysis (GSEA) to identify putative biological process pathways directly affected by DNA methylation. CpG sites that were associated with vitamin intake at FDR ≤ 0.01 were included in gene ontology analysis using the ‘goana’ function in the R package limma [Citation44]. Similarly, we conducted a second gene ontology analysis of all genes that were significantly associated with both vitamin C and E intake at FDR ≤ 0.05 to identify potentially shared pathways. Significant pathways in GSEA were identified at FDR ≤ 0.05.
CpG sites that were significantly associated with vitamin C or E intake at a FDR ≤ 0.01 were used in expression quantitative trait methylation (eQTM) analysis within the FHS cohort to identify downstream effects of DNA methylation on gene expression. eQTM analysis in the FHS has used DNA methylation data and RNA sequencing data to quantify the association between DNA methylation and gene expression independent of age, sex, white blood cell count, blood cell fraction, platelet count, genetic principal components (PCs), and DNA methylation PCs [Citation45]. We identified significant cis associations between CpG sites and gene transcripts (i.e., associations where the CpG site was within 1Mb of the transcription start site) at a p-value of 1.0 × 10−7 [Citation46]. For all significant cis-eQTM CpG-transcript pairs associated with vitamin C or vitamin E intake, we conducted a second GSEA of the gene transcripts to identify pathways that may be affected by vitamin intake-associated DNA methylation.
Exploratory analyses: FHS discovery cohorts
Due to differences in methodology across cohorts, we conducted a series of exploratory and hypothesis-generating analyses in the discovery FHS Offspring and FHS Third-Generation cohorts. All statistical models described in this section were adjusted for all covariates in the primary model: age, sex, BMI, daily caloric intake, blood cell type proportion, smoking status, alcohol consumption, and assay-specific covariates.
Residualized analysis accounting for energy intake
We investigated the association between vitamin and total energy intake in a residual model to evaluate whether our main statistical models were fully accounting for total energy intake through statistical adjustment. Using this approach, we ran an initial regression model to calculate the residuals for the association between log-transformed continuous vitamin intake and total energy, and these residuals were used in a subsequent statistical model to quantify the association between vitamin intake and DNA methylation.
Stratified analyses
To explore potential differences in DNA methylation by demographic, dietary, or behavioural factors, we performed several stratified analyses in the discovery cohort that combined both the FHS Offspring and FHS Third-Generation cohorts. We stratified the associations between DNA methylation and dietary intake of vitamins C and E by 1) male versus female sex; 2) age <65 years versus age ≥65 years; and 3) smoking status (current, former, and never smokers). Specifically, we ran regression models quantifying the adjusted association between DNA methylation and vitamin intake within each stratum of interest and identified CpG sites that were significantly associated with vitamin intake for each stratum at a FDR ≤ 0.05.
Results
Participant characteristics
Characteristics of participants from the study cohort are presented in .
Table 1. Participant characteristics by study cohort.
Vitamin C and DNA methylation
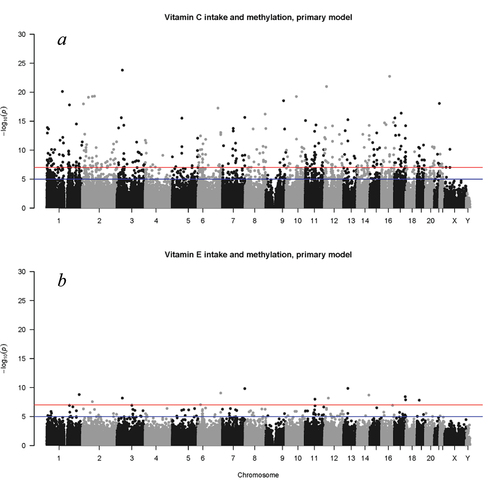
In the meta-analysis combining EWAS results of all 8 cohorts, DNA methylation at 4,656 CpG sites was significantly associated with vitamin C intake after adjusting for age, sex, BMI, caloric intake, blood cell type proportions, smoking status, alcohol consumption, and technical covariates in the primary model (, Supplemental Table S2). These CpG sites were annotated to 2,586 unique genes. Vitamin C intake was associated with hypomethylation at 3,662 (79%) of these CpG sites, and with hypermethylation at the remaining 994 (21%) CpG sites. After further adjustment for diet quality and physical activity, vitamin C intake was associated with hypomethylation at 1,405 CpG sites that were annotated to 986 unique genes, and with hypermethylation at 389 CpG sites annotated to 267 unique genes (Supplemental Table S3).
Figure 2. Manhattan plot of results of the meta-analysis including all study cohorts showing significant associations of Infinium HumanMethylation 450K BeadChip CpG sites with (a) vitamin C intake and (b) vitamin E intake after adjustment for age, sex, BMI, caloric intake, blood cell type proportion, smoking status, alcohol consumption, and technical covariates in the primary model. Blue and red horizontal lines indicate -log(p) values of 5 and 7, respectively (i.e., p = 1.0 × 10−5 and 1.0 × 10−7, representing a less stringent significance threshold for hypothesis generating and a more stringent approximately Bonferroni-corrected threshold, respectively).
In discovery analyses, methylation at 3,643 CpG sites was significantly associated with vitamin C intake in the FHS cohorts after adjustment for all covariates in the primary model, of which 1,521 CpG sites overlapped with the significant results of the overall meta-analysis. Hypermethylation at 17 (0.5%) sites and hypomethylation at 256 (7%) sites was significantly associated with vitamin C intake in the meta-analysis of the replication cohorts at FDR ≤ 0.05 (Supplemental Table S4).
Vitamin E and DNA methylation
DNA methylation at 160 CpG sites was significantly associated with vitamin E intake after adjusting for age, sex, BMI, caloric intake, blood cell type proportions, smoking status, alcohol consumption, and technical covariates in the primary model (, Supplemental Table S5). These CpG sites were annotated to 126 unique genes. Vitamin E intake was associated with hypomethylation at 115 (72%) of these CpG sites, and with hypermethylation at the remaining 45 (28%) CpG sites. After further adjustment for diet quality and physical activity, vitamin E intake was associated with hypermethylation at 17 CpG sites and with hypomethylation at 39 CpG sites (Supplemental Table S6).
In discovery analyses, methylation at 188 CpG sites was significantly associated with vitamin E intake in the FHS cohorts after adjustment for all covariates in the primary model, of which 81 CpG sites overlapped with the significant results of the overall meta-analysis. Hypomethylation of two of the CpG sites (1.1%) and hypermethylation of one of the CpG sites (0.5%) significantly associated with vitamin E intake in discovery analysis was replicated at FDR ≤ 0.05 (Supplemental Table S7).
In the primary model, DNA hypomethylation at 77 of the 160 CpG sites significantly associated with vitamin E intake was also significantly associated with vitamin C intake; similarly, DNA hypermethylation at eight CpG sites significantly associated with vitamin E intake was also significantly associated with vitamin C intake. After adjustment for diet quality and physical activity, 20 CpG sites were significantly associated with both vitamin C and E intake. Of these, 19 CpG sites were hypomethylated in association with both vitamin C and E and annotated to 18 unique genes ().
Table 2. EWAS meta-analysis results for CpG sites at which DNA hypomethylation was associated with both vitamin C and vitamin E intake at FDR ≤ 0.05 after adjusting for age, sex, BMI, caloric intake, blood cell type proportion, smoking status, alcohol consumption, technical covariates, physical activity, and diet.
Downstream pathways affected by DNA methylation: eQTM and GSEA
At FDR ≤ 0.01, there were 1,907 CpG sites significantly associated with vitamin C in the primary model. These CpG sites were annotated to 1,163 unique genes that were significantly enriched in 673 pathways associated with system development and cell signalling and communication (Supplemental Table S8). These 1,907 CpG sites were also associated with expression of 425 unique gene transcripts in eQTM analysis (Supplemental Table S9). The gene transcripts identified by eQTM analysis were enriched in 44 biological pathways that were largely related to the immune response (Supplemental Table S10).
At FDR ≤ 0.01, there were 37 CpG sites that were significantly associated with vitamin E intake in the primary model. These CpG sites were annotated to 29 genes; these genes were not enriched in any cellular or biological pathways. These 37 CpG sites were also associated with the expression of 9 genes (Supplemental Table S11). These genes were not enriched in any biological pathways at an enrichment FDR ≤ 0.05.
Downstream pathways affected by DNA methylation: CpG sites associated with both vitamin C and vitamin E intake
The 19 CpG sites at which DNA methylation was associated with intake of both vitamins C and E were not enriched in any pathways in GSEA.
Exploratory analyses in the discovery FHS cohorts
Residualized analyses
Inclusion of the residualized association between vitamin intake and total energy intake revealed 261 significant associations between continuous vitamin C intake and DNA methylation (Supplemental Table S12), where hypomethylation was associated with vitamin C intake at 227 CpG sites and hypermethylation was associated with vitamin C intake at 34 CpG sites. Similarly, residualized analysis revealed 26 significant associations between continuous vitamin E intake and DNA methylation (Supplemental Table S13), where hypomethylation was associated with vitamin E intake at 23 CpG sites and hypermethylation was associated with vitamin E intake at 3 CpG sites.
Sex-stratified results
In men, DNA hypomethylation at 78 CpG sites was significantly associated with vitamin C intake and DNA hypermethylation at 39 CpG sites was significantly associated with vitamin C intake (Supplemental Table S14). In women, DNA hypomethylation at 40 CpG sites and hypermethylation at 11 CpG sites were associated with vitamin C intake (Supplemental Table S15). Only two CpG sites were associated with vitamin C intake in both men and women: cg03084350 (PLCD1) and cg09018739 (CPNE2).
In men, there were no significant associations between DNA methylation and vitamin E intake; however, in women, DNA hypomethylation at three CpG sites was significantly associated with vitamin E intake (Supplemental Table S16).
Age-stratified results
In participants aged 65 years and older, DNA hypomethylation at 32 CpG sites and hypermethylation at 24 CpG sites was associated with vitamin C intake (Supplemental Table S17). In participants under the age of 65 years, DNA hypomethylation at 73 CpG sites and hypermethylation at 11 CpG sites was associated with vitamin C intake (Supplemental Table S18). Four CpG sites were associated with vitamin C intake in both age strata.
In participants aged 65 years and older, DNA hypomethylation at two CpG sites and hyper-methylation at one CpG site were associated with vitamin E intake (Supplemental Table S19). In participants under the age of 65 years, DNA hypomethylation at six CpG sites and hypermethylation at one CpG site were associated with vitamin E intake (Supplemental Table S20). No CpG sites were associated with vitamin E intake in both age strata.
Smoking status-stratified results
In never smokers, DNA hypomethylation at seven CpG sites and hypermethylation at 11 CpG sites were associated with vitamin C intake (Supplemental Table S21). In former smokers, DNA hypomethylation at 64 CpG sites and hypermethylation at 16 CpG sites were associated with vitamin C intake (Supplemental Table S22). Finally, in current smokers, DNA hypomethylation at four CpG sites and hypermethylation at five CpG sites were associated with vitamin C intake (Supplemental Table S23).
DNA methylation was not significantly associated with vitamin E intake at any CpG sites in never or in current smokers. DNA hypomethylation was significantly associated with vitamin E intake at one CpG site only – cg13574809 – in former smokers (Supplemental Table S24).
Discussion
In this study, we combined data across multiple large population-based cohorts to examine the association of vitamin C and E intake with DNA methylation signatures. We provide evidence that greater intake of vitamins C and E is associated with DNA methylation signatures, and identified several thousand CpG sites associated with vitamin C intake, and approximately 100 CpG sites associated with vitamin E intake. The majority of these sites were hypomethylated in relation to increased vitamin intake, even in stratified analysis; furthermore, DNA methylation at the CpG sites identified in EWAS was inversely associated with gene expression in subsequent eQTM analysis.
Oxidative stress affects cellular and biological processes, and it can lead to inflammation and development of clinical disease, including atherosclerosis and metabolic disease [Citation47–50]. Antioxidants from a healthy diet, supplementation, or pharmacological interventions may improve immune response and prevent oxidative stress-related cardiometabolic disease. In humans, vitamin C must be obtained exogenously either through diet (e.g., citrus fruit, cruciferous vegetables, tomatoes), supplementation, or pharmacological administration. It acts as a cofactor of TET proteins, which catalyse the hydroxylation of methyl groups at the cytosine C5 position for subsequent removal in DNA demethylation. Conversely, vitamin E is a lipid-soluble antioxidant found in cell membranes that interrupts radical chain reactions and may subsequently prevent inflammation [Citation21,Citation51,Citation52]. It interacts with vitamin C to subsequently become reduced back into active vitamin E and then continue acting as an antioxidant in the cell membrane. Similar to vitamin C, it must be obtained through exogenous means (e.g., consumption of seeds, nuts, and oils). Prior studies have suggested a role of vitamin intake-associated DNA methylation in the expression of particular genes, such as the ABCA1 gene [Citation53] or genes associated with oxidative stress [Citation54] We identified 85 CpG sites at which DNA methylation was significantly associated with both vitamin C and vitamin E intake, of which 20 CpG sites had an association that was independent of other lifestyle or behavioural risk factors. Although enrichment analysis did not reveal a pathway in which the genes annotated to these CpG sites DNA methylation at these sites may play a role, this overlap is suggestive of the known antioxidative interaction between vitamins C and E. Future studies may evaluate the cellular and biological effects of suppressing the genes associated with intake of both vitamins, as our results showed predominant associations of increased dietary vitamin intake with DNA hypomethylation.
While the observed differences in our stratified analyses were limited by reduced power in the discovery cohort, we generally saw that there were differences in which CpG sites were associated with vitamin intake by age, sex, and smoking status, particularly in association with vitamin C intake. In summary, men, individuals less than 65 years of age, and former smokers had the greatest number of significant associations between DNA methylation and vitamin C intake as compared with women, individuals over 65 years of age, never smokers, and current smokers. Like the global meta-analysis results, vitamin C intake was mostly associated with DNA hypomethylation among all stratified groups: hypomethylation at 67%, 87%, and 80% of CpG sites significantly associated with vitamin C intake in men, individuals younger than 65 years, and former smokers, respectively. These preliminary results may reflect a more active role for vitamin C in the setting of higher oxidative stress (i.e., in men and in former smokers) or without accounting for other behaviours potentially found in adults younger than 65 years. Further studies may focus on expanding these stratified analyses to larger cohorts.
Our results further suggest a role for vitamin C-associated DNA methylation patterns in activation of the immune response. We identified numerous biological pathways potentially directly affected by vitamin C-associated methylation: response to stimulus, regulation of biological and cellular processes, and cell signalling and communication. Further analysis of eQTM CpG-gene transcript pairs showed that the DNA methylation patterns associated with vitamin C intake are associated with the downstream expression of genes that are in the immune response pathway. Vitamin E has also previously been shown to be associated with the inflammatory response [Citation55–57]; however, while prior studies have shown an association between vitamin E concentration and expression of the DNMT1 gene [Citation58,Citation59], we were not able to identify any gene ontology pathways associated with vitamin E-associated DNA methylation.
While this study benefits from the availability of data from large population-based cohorts to evaluate the association of vitamin intake with both DNA methylation and gene expression, several limitations must be considered when interpreting and applying our results. Firstly, as these data were collected as part of large observational studies, causal inference is not possible; future mediation analyses evaluating the relationship between dietary vitamin intake, DNA methylation, and gene expression will reveal potential causal mechanisms. Our meta-analysis included only individuals with European and African ancestry, and generalizability of these findings to other populations may be limited. While the majority of participants included in our meta-analysis were adults, the 512 participants enrolled in the Raine Study (4% of the total participants included in the main meta-analysis) were 17 years of age at the time of data collection. Individuals below the age of 18 have different vitamin C and E recommended dietary allowance thresholds as compared with adults over the age of 18. Additionally, when we split the overall sample for our main analysis into a discovery cohort and a replication cohort, we observed poor replication for both the vitamin C and the vitamin E analyses, potentially due to major differences in how these FFQs captured and reported vitamin intake and consequent high heterogeneity across cohorts for some CpG sites. Dietary intake of vitamins C and E was based solely on self-reported dietary intake rather than on a biomarker measure, and we did not include analysis of any other antioxidants in our analysis, preventing analysis of their potential effects on DNA methylation. We also used categorical vitamin intake based on clinically relevant thresholds in our main analyses, although we completed sensitivity analyses that residualized total energy intake by continuous vitamin intake in our discovery cohort. However, due to non-normal distributions of self-reported vitamin intake, our approach allows for easier interpretation of the association between levels of vitamin intake and DNA methylation, and it is consistent across all included cohorts. Our definition of vitamin C intake included the use of supplements, and supplemental intake of multivitamins may introduce confounding due to the presence of folic acid and other nutrients that are associated with DNA methylation. Prior analyses have shown few significant associations between DNA methylation and folate intake [Citation60,Citation61], and so we expect these nutrients not to influence our observed results greatly. While we adjusted for diet quality using the Mediterranean diet score to account for overall healthy diets, simultaneous supplemental intake of multiple vitamins and nutrients may not be adequately captured. Furthermore, while our secondary model did adjust for lifestyle and behavioural risk factors by accounting for diet quality and physical activity level, we did not have data on and thus could not account for environmental exposures that are known to be associated with DNA methylation patterns.
In conclusion, dietary and supplementary intake of vitamin C and vitamin E is associated with DNA methylation, which likely plays a critical role in the regulation of cell signalling mechanisms and subsequently, in the association of vitamin intake with the immune response. Our results highlight the importance of dietary and supplementary intake of vitamins C and E through consumption of fruit, vegetables, nuts, seeds, and oils in overall health and the reduction of burden from inflammatory processes.
Disclaimer
The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services.
Data availability
FHS: Data from the FHS Offspring and Third-Generation cohorts are available at the Database of Genotypes and Phenotypes (dbGaP) at https://www.ncbi.nlm.nih.gov/gap/, reference number phs000007.v29.p10.
TwinsUK: Many of the data analysed in the current study is available through GEO GSE62992 and GSE121633. Additional individual-level data are not permitted to be shared or deposited due to the original consent given at the time of data collection. However, access to these data can be applied for through the TwinsUK data access committee. For information on access and how to apply http://twinsuk.ac.uk/resources-for-researchers/access-our-data/.
KORA: The informed consents given by KORA study participants do not cover data posting in public databases. However, data are available upon request from KORA Project Application Self-Service Tool (https://helmholtz-muenchen.managed-otrs.com/external) Data requests can be submitted online and are subject to approval by the KORA Board.
Young Finns Study: The dataset supporting the conclusions of this article were obtained from the Cardiovascular Risk In Young Finns study which comprises health-related participant data. The use of data is restricted under the regulations on professional secrecy (Act on the Openness of Government Activities, 612/1999) and on sensitive personal data (Personal Data Act, 523/1999, implementing the EU data protection directive 95/46/EC). Due to these restrictions, the data can not be stored in public repositories or otherwise made publicly available. Data access may be permitted on a case by case basis upon request only. Data sharing outside the group is done in collaboration with YFS group and requires a data-sharing agreement. Investigators can submit an expression of interest to the chairman of the publication committee (Prof Mika Kähönen, Tampere University, Finland) or Professor Terho Lehtimäki (Tampere University, Finland) concerning the epigenetic and genetic data.
Raine Study: The Raine Study holds a rich and detailed collection of data gathered over 30 years for the purpose of health and well-being research. The informed consent provided by each participant does not permit individual-level data to be made available in the public domain (i.e., a public data repository). However, de-identified analytic data sets are available to all researchers for original research or auditing of published findings. All data access is managed through established Raine Study procedures which require data handlers to agree to a code of conduct that includes safeguards to protect the identity of participants. Details of the data access processes, and code of conduct are available on the Raine Study website https://rainestudy.org.au/. The governance process for accessing the Raine Study data can be accessed here: https://rainestudy.org.au/information-for-researchers/information-for-new-researchers/.
Rotterdam Study: The Rotterdam Study data can be made available to interested researchers upon reasonable request. Requests can be directed to data manager Frank J.A. van Rooij ([email protected]). We are unable to place data in a public repository due to legal and ethical restraints. Sharing of individual participant data was not included in the informed consent of the study, and there is potential risk of revealing participants’ identities as it is not possible to completely anonymize the data. This is of particular concern given the sensitive personal nature of much of the data collected as part of the Rotterdam Study.
ARIC Study: The DNA methylation dataset from ARIC is available upon request at https://sites.cscc.unc.edu/aric/distribution-agreements.
Supplemental Material
Download Zip (3 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592294.2023.2211361
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- McMahon KW, Karunasena E, Ahuja N. The roles of DNA methylation in the stages of cancer. Cancer J. 2017;23(5):257–15.
- Severi G, Southey MC, English DR, et al. Epigenome-wide methylation in DNA from peripheral blood as a marker of risk for breast cancer. Breast Cancer Res Treat. 2014;148(3):665–673.
- Battram T, Richmond RC, Baglietto L, et al. Appraising the causal relevance of DNA methylation for risk of lung cancer. Int J Epidemiol. 2019;48(5):1493–1504.
- Ahmed SAH, Ansari SA, Mensah-Brown EPK, et al. The role of DNA methylation in the pathogenesis of type 2 diabetes mellitus. Clin Epigenetics. 2020;12(1):104.
- Liu J, Carnero-Montoro E, van Dongen J, et al. An integrative cross-omics analysis of DNA methylation sites of glucose and insulin homeostasis. Nat Commun. 2019;10(1):2581.
- Richard MA, Huan T, Ligthart S, et al. DNA methylation analysis identifies loci for blood pressure regulation. Am J Hum Genet. 2017;101(6):888–902.
- Zawia NH, Lahiri DK, Cardozo-Pelaez F. Epigenetics, oxidative stress, and Alzheimer disease. Free Radic Biol Med. 2009;46(9):1241–1249.
- Franco R, Schoneveld O, Georgakilas AG, et al. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. 2008;266(1):6–11.
- Hedman ÅK, Zilmer M, Sundström J, et al. DNA methylation patterns associated with oxidative stress in an ageing population. BMC Med Genomics. 2016;9(1):72.
- Donkena KV, Young CYF, Tindall DJ. Oxidative stress and DNA methylation in prostate cancer. Obstet Gynecol Int. 2010;2010:302051.
- Włodarczyk M, Jabłonowska-Lietz B, Olejarz W, et al. Anthropometric and dietary factors as predictors of DNA damage in obese women. Nutrients. 2018;10:578.
- Salonen RM, Nyyssönen K, Kaikkonen J, et al. Six-year effect of combined vitamin C and E supplementation on atherosclerotic progression: the antioxidant supplementation in atherosclerosis prevention (ASAP) study. Circulation. 2003;107(7):947–953.
- Lutsenko EA, Cárcamo JM, Golde DW. Vitamin C Prevents DNA Mutation Induced by Oxidative Stress *. J Biol Chem. 2002;277(19):16895–16899.
- Attri J, Dhawan V, Mahmood S, et al. Effect of Vitamin C supplementation on oxidative dna damage in an experimental model of lead-induced hypertension. Ann Nutr Metab. 2003;47(6):294–301.
- Kawashima A, Sekizawa A, Koide K, et al. Vitamin C induces the reduction of oxidative stress and paradoxically stimulates the apoptotic gene expression in extravillous trophoblasts derived from first-trimester tissue. Reprod Sci. 2015;22(7):783–790.
- Blaschke K, Ebata KT, Karimi MM, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–226.
- Lee Chong T, Ahearn EL, Cimmino L. Reprogramming the epigenome with vitamin C. Front Cell Dev Biol. 2019;7:128.
- Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935.
- Beetch M, Harandi-Zadeh S, Shen K, et al. Dietary antioxidants remodel DNA methylation patterns in chronic disease. Br J Pharmacol. 2020;177(6):1382–1408.
- Niki E. Interaction of ascorbate and α-Tocopherol. Ann N Y Acad Sci. 1987;498(1 Third Confere):186–199.
- Traber MG, Stevens JF. Vitamins C and E: beneficial effects from a mechanistic perspective. Free Radic Biol Med. 2011;51(5):1000–1013.
- Thomas SR, Stocker R. Molecular action of vitamin E in lipoprotein oxidation: implications for atherosclerosis. Free Radic Biol Med. 2000;28(12):1795–1805.
- Kagan VE, Serbinova EA, Forte T, et al. Recycling of vitamin E in human low density lipoproteins. J Lipid Res. 1992;33(3):385–397.
- Kannel WB, Feinleib M, McNamara PM, et al. An investigation of coronary heart disease in families: the framinham offspring study. Am J Epidemiol. 1979;110(3):281–290.
- Splansky GL, Corey D, Yang Q, et al. The third generation cohort of the national heart, lung, and blood institute’s framingham heart study: design, recruitment, and initial examination. Am J Epidemiol. 2007;165(11):1328–1335.
- Moayyeri A, Hammond CJ, Valdes AM, et al. Cohort Profile: TwinsUK and healthy ageing twin study. Int J Epidemiol. 2013;42(1):76–85.
- Holle R, Happich M, Löwel H, et al. Kora–a research platform for population based health research. Gesundheitswes (Bundesverband der Arzte des Offentl Gesundheitsdienstes. 2005;67(1):S19–25
- Kowall B, Rathmann W, Stang A, et al. Perceived risk of diabetes seriously underestimates actual diabetes risk: the KORA FF4 study. PLoS ONE. 2017;12:e0171152.
- Akerblom HK, Viikari J, Uhari M, et al. Atherosclerosis precursors in Finnish children and adolescents. I. General description of the cross-sectional study of 1980, and an account of the children’s and families’ state of health. Acta Paediatr Scand Suppl. 1985;318(s318):49–63.
- Straker L, Mountain J, Jacques A, et al. Cohort profile: the western australian pregnancy cohort (raine) study–generation 2. Int J Epidemiol. 2017;46:1384–1385j.
- Ikram MA, Brusselle G, Ghanbari M, et al. Objectives, design and main findings until 2020 from the rotterdam study. Eur J Epidemiol. 2020;35(5):483–517.
- The ARIC Investigators. The atherosclerosis risk in communities (ARIC) study: design and objectives. Am J Epidemiol. 1989;129(4):687–702. DOI:10.1093/oxfordjournals.aje.a115184
- Willett WC, Sampson L, Stampfer MJ, et al. Reproducibility and validity of a semiquantitative food frequency questionnaire. Am J Epidemiol. 1985;122(1):51–65.
- Bingham SA, Welch AA, McTaggart A, et al. Nutritional methods in the European prospective investigation of cancer in norfolk. Public Health Nutr. 2001;4(3):847–858.
- Mitry P, Wawro N, Six-Merker J, et al. Usual dietary intake estimation based on a combination of repeated 24-h food lists and a food frequency questionnaire in the KORA FF4 cross-sectional study. Front Nutr. 2019;6:145.
- Feskanich D, Rimm EB, Giovannucci EL, et al. Reproducibility and validity of food intake measurements from a semiquantitative food frequency questionnaire. J Am Diet Assoc. 1993;93(7):790–796.
- Ambrosini GL, de Klerk NH, O’Sullivan TA, et al. The reliability of a food frequency questionnaire for use among adolescents. Eur J Clin Nutr. 2009;63(10):1251–1259.
- Voortman T, Jong JC KD, Ikram MA, et al. Adherence to the 2015 Dutch dietary guidelines and risk of non-communicable diseases and mortality in the Rotterdam Study. Eur J Epidemiol. 2017;32(11):993–1005.
- Stevens J, Metcalf PA, Dennis BH, et al. Reliability of a food frequency questionnaire by ethnicity, gender, age and education. Nutr Res. 1996;16(5):735–745.
- Monsen ER. Dietary Reference Intakes for the Antioxidant Nutrients: vitamin C, Vitamin E, Selenium, and Carotenoids. J Am Diet Assoc. 2000;100(6):637–640.
- Sofi F, Macchi C, Abbate R, et al. Mediterranean diet and health status: an updated meta-analysis and a proposal for a literature-based adherence score. Public Health Nutr. 2014;17(12):2769–2782.
- Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 2012;13:86.
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B. 1995;57(1):289–300.
- Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47.
- Keshawarz A, Bui H, Joehanes R, et al. Expression quantitative trait methylation analysis elucidates gene regulatory effects of DNA methylation: the Framingham Heart Study. medRxiv. 2022;2022.04.13.22273839.
- Yao C, Joehanes R, Wilson R, et al. Epigenome-wide association study of whole blood gene expression in Framingham Heart Study participants provides molecular insight into the potential role of CHRNA5 in cigarette smoking-related lung diseases. Clin Epigenetics. 2021;13(1):60.
- Tangvarasittichai S. Oxidative stress, insulin resistance, dyslipidemia and type 2 diabetes mellitus. World J Diabetes. 2015;6(3):456–480.
- Yoshida H, Kisugi R. Mechanisms of LDL oxidation. Clin Chim Acta. 2010;411(23–24):1875–1882.
- Jha JC, Ho F, Dan C, et al. A causal link between oxidative stress and inflammation in cardiovascular and renal complications of diabetes. Clin Sci (Lond). 2018;132(16):1811–1836.
- Garcia-Bailo B, El-Sohemy A, Haddad PS, et al. Vitamins D, C, and E in the prevention of type 2 diabetes mellitus: modulation of inflammation and oxidative stress. Biologics. 2011;5:7–19.
- van Dam B, van Hinsbergh VWM, Stehouwer CDA, et al. Vitamin E inhibits lipid peroxidation-induced adhesion molecule expression in endothelial cells and decreases soluble cell adhesion molecules in healthy subjects. Cardiovasc Res. 2003;57(2):563–571.
- Reaven PD, Herold DA, Barnett J, et al. Effects of Vitamin Eon susceptibility of low-density lipoprotein and low-density lipoprotein subfractions to oxidation and on protein glycation in NIDDM. Diabetes Care. 1995;18:807–816.
- Fujii R, Yamada H, Munetsuna E, et al. Associations between dietary vitamin intake, ABCA1 gene promoter DNA methylation, and lipid profiles in a Japanese population. Am J Clin Nutr. 2019;110(5):1213–1219.
- Shorey-Kendrick LE, McEvoy CT, O’Sullivan SM, et al. Impact of vitamin C supplementation on placental DNA methylation changes related to maternal smoking: association with gene expression and respiratory outcomes. Clin Epigenetics. 2021;13(1):177.
- Meydani SN, Meydani M, Blumberg JB, et al. Vitamin E supplementation and in vivo immune response in healthy elderly subjects. A randomized controlled trial. JAMA. 1997;277(17):1380–1386.
- Meydani SN, Meydani M, Rall LC, et al. Assessment of the safety of high-dose, short-term supplementation with vitamin E in healthy older adults. Am J Clin Nutr. 1994;60(5):704–709.
- Meydani SN, Han SN, Wu D. Vitamin E and immune response in the aged: molecular mechanisms and clinical implications. Immunol Rev. 2005;205(1):269–284.
- Remely M, Ferk F, Sterneder S, et al. Vitamin E Modifies High-Fat Diet-Induced Increase of DNA Strand Breaks, and Changes in Expression and DNA Methylation of Dnmt1 and MLH1 in C57BL/6J Male Mice. Nutrients. 2017;9:607.
- Zappe K, Pointner A, Switzeny OJ, et al. Counteraction of Oxidative Stress by Vitamin E Affects Epigenetic Regulation by Increasing Global Methylation and Gene Expression of MLH1 and DNMT1 Dose Dependently in Caco-2 Cells. Oxid Med Cell Longev. 2018;2018:3734250.
- Mandaviya PR, Joehanes R, Brody J, et al. Association of dietary folate and vitamin B-12 intake with genome-wide DNA methylation in blood: a large-scale epigenome-wide association analysis in 5841 individuals. Am J Clin Nutr. 2019;110(2):437–450.
- Kok DEG, Dhonukshe-Rutten RAM, Lute C, et al. The effects of long-term daily folic acid and vitamin B12 supplementation on genome-wide DNA methylation in elderly subjects. Clin Epigenetics. 2015;7(1):121.