ABSTRACT
The histone H3K27 demethylase, UTX/KDM6A, plays a critical role in the early development of vertebrates, and mutations are frequently found in various cancers. Several studies on developmental and cancer biology have focused on preferential transcriptional regulation by UTX independently of its H3K27 demethylase catalytic activity. Here, we analysed gene expression profiles of wild-type (WT) UTX and a catalytic activity-defective mutant in 786-O and HCT116 cells and confirmed that catalytic activity-dependent and -independent regulation contributes to the expression of most of the target genes. Indeed, the catalytic activity-defective mutant suppressed colony formation similar to the WT in our assay system. However, the expression of several genes was significantly dependent on the catalytic activity of UTX in a cell type-specific manner, which could account for the inherent variation in the transcriptional landscape of various cancer types. The promoter/enhancer regions of the catalytic activity-dependent genes identified here were found to be preferentially modified with H3K4me1 and less with H3K27me3 than those of the independent genes. These findings, combined with previous reports, highlight not only the understanding of determinants for the catalytic activity dependency but also the development and application of pharmaceutical agents targeting the H3K27 or H3K4 modifications.
Introduction
UTX, also referred to as KDM6A, is one of the major histone H3K27 demethylases [Citation1–4] and a component of MLL3/4-containing multi-protein complexes [Citation5–7]. Mechanistically, it antagonizes the histone H3K27 methyltransferase complex PRC2 containing the catalytic subunit EZH2 [Citation8–10]. UTX has paralogs, the catalytically active JMJD3 (KDM6B) with a moderate conservation of amino acid identity and the Y-linked UTY with even higher conservation but null or very low in the catalytic activity [Citation1,Citation4]. While the catalytic activity of histone modifiers has generally been believed to be critical for their function, the importance of catalytic activity-independent UTX functions has also been documented for both early development and cancer formation. Notably, significant differences between UTX and UTY functions, which highlight the catalytic activity-dependent aspect of UTX, have also been observed.
Utx null female mice (Utx-/-) display severe developmental defects mainly in mesodermal organs and fail to survive after mid-gestation, whereas Utx-deficient male mice Utx−/Uty+ develop with normal morphology until mid-gestation and survive until the perinatal stage or even longer after birth [Citation11–13]. Male mice with further disruption of Uty (Utx−/Uty−) phenocopy the Utx-/- female, suggesting that Uty can partially compensate for loss of UTX function. In addition, UTX catalytic activity-disrupted male ES cells (UtxKI/Uty+) generated by knock-in experiments can differentiate into mesoderm-like cells [Citation12]. The phenotypic severity of Utx−/Uty+ mice has been variably reported: Utx−/Uty+ phenotypes are closer to that of the wild-type in some cases [Citation13] or moderately to severely affected with manifestations of neural crest closure defects, cardiac malformation, and tumour formation in adults [Citation11,Citation12]. This suggests that the UTX catalytic activity, which has been evolutionarily conserved, is required for some processes of development. Target genes of UTX include cardiac-specific transcription factors such as Srf, Nkx2.5, Gata4, and Tbx5 in the heart [Citation11], development-related genes such as Brachyury, Fgf5, Hmga2, Pitx2, Wnt3, and Wt1 in ES cells [Citation12], and Fnbp1 in MEF cells [Citation13]. Many of these target genes are regulated in a UTX catalytic activity-independent manner yet to variable extents [Citation12,Citation13]. These results highlight the critical involvement of catalytic activity-independent and cell type-specific functions of Utx in development.
Based on extensive large-scale sequencing data, aetiological evidence for UTX as a tumour suppressor has been increasing. UTX mutations have been found in various human cancer types, with large variation in frequency, including in clear cell renal cell carcinoma, medulloblastoma, chronic myelomonocytic leukaemia, acute lymphoblastic leukaemia, transitional cell carcinoma of the bladder, and high-grade muscle-invasive urothelial bladder carcinoma [Citation14,Citation15]. However, Utx heterozygous or homozygous knockout (KO) mice alone do not develop malignant lesions [Citation11–13]. This is likely because UTX requires concurrent alterations in other genes to develop full malignancy [Citation16]. Consistently, Utx disruption in cancer-prone backgrounds (e.g., with exogenous expression of activated Notch, c-Myc, or KrasG12D) in mice leads to malignancy of haematopoietic cells and some other tissues [Citation17–20]. Andricovich et al. reported that gender-specific malignancies develop by pancreas-targeted Utx disruption in mice with KrasG12D background [Citation19]. In this report, Utx-/Y mice showed intermediate but closer to the wild type (WT) phenotypes in the Kaplan–Meier plots and histology. Remarkably, Gozdecka et al. showed that homozygous loss of Utx in mice with no additional genetic alterations efficiently confers a preleukemic state in the spleen and bone marrow, which was suppressed by the presence of Uty (Utx−/Uty+) [Citation21]. This suggests that loss of Utx can lead to development of the haematopoietic malignancies in a catalytic activity-independent manner. Shi et al. found that the core intrinsically disordered region (cIDR) as well as the amino-terminal tetratricopeptide repeat (TPR) region are indispensable for phase-separated condensation of UTX in nuclear foci and xenograft formation in mice [Citation22]. It was further shown that UTY had a stronger condensation ability than UTX and suggested that this could account for the weaker tumour suppressor activity of UTY. As such, also in cancer formation of mice, Uty compensation for Utx loss as a tumour suppressor is undoubted, but the extent has been variably described. Paradoxically, UTX knockdown or disruption attenuates cell proliferation, colony formation, and xenograft growth of breast cancer cells [Citation23,Citation24], and UTX copy loss and gain in bladder carcinoma are neutral (20% and 15%, respectively, Cansar). Furthermore, the H3K27 methyltransferase EZH2, a presumed oncogene with a possible antagonistic activity to UTX, also showed unexpected tumour suppressive properties in Kras-driven lung adenocarcinoma and diffuse midline glioma [Citation25,Citation26]. Taken together, although UTX functions as a tumour suppressor in various cell types and tissues, UTX involvement in cancer formation appears to be cell type-, tissue- and context-dependent and the underlying mechanisms have largely remained to be elucidated [Citation27].
The requirement of the UTX catalytic activity in naturally occurring cancer cells has also been addressed with cultured cells. Overexpression of WT UTX in UTX-null cell lines, but not in controls, results in cell growth retardation [Citation28], whereas overexpression of catalytic activity-defective mutants in a WT line increases cell growth [Citation29]. This suggests the involvement of UTX catalytic activity in tumour growth suppression. However, in a more focused study, a catalytic activity-defective UTX (HEAA UTX) expressed in a UTX-null bladder carcinoma line suppressed colony formation, activated luminal marker genes, and localized on their promoters, all nearly comparably to WT [Citation30]. However, the extent to which UTX target genes (i.e., UPK1B, UPK2, and UPK3B) were upregulated by HEAA UTX was consistently and significantly lower than that driven by WT [Citation30]. We previously showed that UTX interacts with MLL3/4 complexes through the UTX TPR region and that a cancer-derived UTX defective in its interaction with MLL3/4 complexes relocates to the cytoplasm, and introduction of WT UTX but not this mutant suppresses colony formation of UTX-negative cancer cells, arguing for more critical involvement of the interaction than catalytic activity per se [Citation31]. Mechanistically, the histone modification activities of MLL3/4 and p300 that are recruited to enhancers by UTX appear to be critical for catalytic activity-independent transcriptional regulation by UTX [Citation32].
Here, we have established and utilized cells stably expressing WT or a catalytic activity-defective UTX (HQE-AQA) in UTX-disrupted clones from 786-O and HCT116 cells and compared their gene expression profiles. 786-O and HCT116 showed mutually distinct UTX target specificities. Most of these genes were regulated largely by the catalytic activity-independent action of UTX to variable extents and in a cell-type-specific manner. Catalytic activity-dependent regulation is evident in some target genes, which could be explained by the difference in epigenetic modification status around the promoter regions. The HQE-AQA mutant suppressed colony formation in soft agar as efficiently as WT, whereas the interaction-defective UTX failed to do so. These results indicate the critical but partial involvement of the catalytic activity-independent function of UTX in gene expression and potentially in cancer formation.
Materials and Methods
Plasmids
WT and mutant UTX proteins were stably expressed with a lentivirus vector (CSII-CMV-IRES2-puro, RIKEN) as previously described [Citation31]. sgRNAs targeting UTX exon 1 in CRISPR-Cas9 vector (lentiCRISPR v2) were used for gene disruption [Citation33,Citation34].
Cell culture and preparation of UTX-disrupted clones by CRISPR-Cas9 gene editing
HCT116, 786-O (ATCC) and their derivatives were cultured in DMEM-high glucose supplemented with 10% foetal bovine serum (FBS). To establish clones with disrupted UTX, sgRNA-lentiCRISPR v2 was transiently transfected and selected with puromycin for 3 d, and single cell-derived colonies were isolated. Genomic sequence and expression of UTX were analysed by genomic PCR followed by direct sequencing and western blotting [Citation34].
Antibodies and western blot analysis
α-UTX (GTX121246) was purchased from GeneTex and α-βactin (A5441) from Sigma-Aldrich. Western blot analysis was performed with the ECL kit (GE Healthcare) as previously described [Citation35].
Gene expression profiling with microarray and bioinformatics data processing
Total RNA was extracted from cell cultures in biological triplicates using the RNeasy Mini kit (Qiagen), and RNA quality was assessed on the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) using the RNA 6000 Nano Chip kit (Agilent) for intact 18S and 28S ribosomal peaks with RNA Integrity Number (RIN) >7.5. Four hundred nanograms of total RNA was reverse-transcribed into cDNA and in vitro transcribed into biotin-labelled cRNA using the TotalPrep RNA Amplification Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s recommendations. Seven hundred and fifty nanograms of the biotin-labelled cRNA from each sample was hybridized to HumanHT-12 v4.0 beadchip microarrays (Illumina) and scanned on the BeadArray Reader (Illumina) at scan factor 1. Raw intensity values were background subtracted using the BeadStudio Data Analysis software 3.1.3.0 software (Illumina; https://www.illumina.com/Documents/products/datasheets/datasheet_beadstudio.pdf) and normalized using the cross-correlation method. Differential gene expression was identified based on a fold change cut-off of 1.5 between the UTX (WT and HQE-AQA)-expressing vector and the empty vector in the CRISPR clones CR1 and CR2 of 786-O and HCT116 cell lines. The gene expression profiling data has been deposited in the Gene Expression Omnibus (GEO, NCBI) with the accession number GSE127860. Cancer dependency and gene expression correlation analyses were performed using DepMap and Morpheus, respectively (Broad Institute, USA). Data for UTX copy loss and gain in bladder cancer were obtained in Cansar (https://cansar.ai/target/O15550/disease/bladder-cancer). The chromatin immunoprecipitation (ChIP) database (ChIP-Atlas) [Citation36] was used to analyse histone modification status in the promoter/enhancer regions (−5 kb to +5 kb from the start sites) of the catalytic activity-dependent and -independent UTX target genes against reference gene promoters (RefSeq) of all cell types with a threshold of significance at 50.
Soft agar assay
Colony formation assay in soft agar layer was previously described [Citation31]. Briefly, 3000 HeLa cells in DMEM-high glucose containing 10% FBS and 0.35% agarose were poured on the bottom layer medium with 0.5% agarose. In 2 weeks, colonies were counted after staining with 0.5% crystal violet. Cell colonies were counted with the Colony Counter plugin of ImageJ (NIH, USA).
RT-qPCR
RT-qPCR protocol was previously described [Citation34,Citation35]. Briefly, total RNA extracted with the RNeasy Mini Kit (Qiagen) was used for cDNA synthesis using the iScript cDNA Synthesis Kit (Bio-Rad, 170–8891). cDNA samples were amplified using the KAPA SYBR FAST qPCR Kit (Kapa Biosystems, KK4602) and quantified by the 7900HT Fast Real-Time PCR system (Applied Biosystems). Primers for PCK2, CHOP, REDD1, CHAC1, and TRIB3 were previously shown [Citation34], while other primers used in this study are indicated below. Biological triplicates (N = 3) were analysed for each sample. Differences between WT and control (empty vector) are statistically significant (p < 0.01) except for cases indicated by n.s./c (not significant to control). Significant differences between WT and HQE-AQA are indicated by asterisks.
EMP1 F: TTGCTGGCTGGTATCTTT, EMP1 R: TTGAGGGCATCTTCACTG
MMP7 F: AGATGTGGAGTGCCAGATGT, MMP7 R: TAGACTGCTACCATCCGTCC
PPIA F: ACGGCGAGCCCTTGG, PPIA R: TTTCTGCTGTCTTTGGGACCT
Statistics
Unpaired two-tailed Student’s t-tests were utilized to calculate p-values (*: p < 0.05; **:p < 0.01). All the microarray and RT-qPCR data (WT and HQE-AQA UTX) were compared against the mean value of controls (empty vector). Error bars were not included for a clearer view.
Results
UTX H3K27 demethylase-dependent and -independent transcriptional regulation in UTX-disrupted 786-O clones
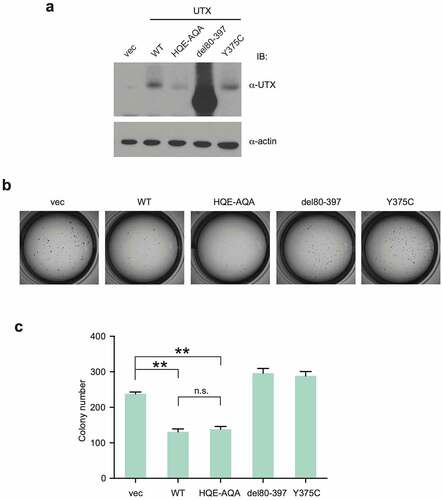
In our previous work, re-expression of UTX in UTX-disrupted HCT116 changes a small number of genes [Citation34]. UTX is one of the genes frequently mutated in clear cell renal cell carcinoma (ccRCC) [Citation28,Citation37] and therefore to investigate UTX transcriptional regulation further, we used a ccRCC line 786-O for disruption and re-expression of UTX in this study. Two independent clones (786-O CR1 and CR2) carrying a small frame-shift mutation in the first exon of UTX were isolated after CRISPR-Cas9 gene editing. Then, WT UTX or UTX with the HQE-AQA catalytic domain mutation was re-expressed by stable introduction with lentivirus vectors (), which can avoid a bias from clonal selection. These cell lines derived from the CR1 and CR2 clones were used for microarray gene expression profiling. Genes upregulated by >1.5 fold in WT-expressing cells, compared to control (empty vector) for both CR1 and CR2 were first identified and aligned with the corresponding data for HQE-AQA (, S1a). Most of the genes display similar upregulation trends between WT and HQE-AQA. However, their expression levels in HQE-AQA were consistently lower, although still higher than those of the control vector level (=1.0) in most cases (). The activation ability was substantially attenuated in HQE-AQA (e.g., EMP1: 45% reduction in CR2; TRIML2: 50% reduction in CR1; ANGPTL4: 48% reduction in CR2; SCARNA18: 100% reduction in CR1) and the differences were statistically significant (p < 0.05 or p < 0.01, ). Notably, transcriptional upregulation of most of these genes only partially depends on the H3K27 demethylase activity of UTX to variable extents. Although the correlation plots show a good association between WT and HQE-AQA UTX re-expression for many target genes, some of these genes deviated from the 1:1 trendline (Figure S2a,b). RT-qPCR analysis was performed for EMP1 (Epithelial Membrane Protein 1), which is the most strongly activated for both CR1 and CR2 () by UTX re-expression. The result closely reproduces the microarray data, suggesting that the quantitation by gene expression profiling with microarrays was reliable. To validate the clinical relevance of this finding, we performed co-expression analysis using Morpheus and found that expression of EMP1 positively correlates with UTX expression in many but not all patient tissue samples including kidney renal clear cell carcinoma (KIRC) and colon adenocarcinoma (COAD) with Pearson correlation values of 0.24 and 0.29, respectively (Figure S3).
Figure 1. Expression profiles of upregulated genes with re-expression of WT or a catalytic activity-defective (HQE-AQA) UTX in UTX-disrupted 786-O cells. (a) WT or HQE-AQA UTX was stably expressed in two 786-O clones with frame-shifted UTX gene (CR1 and CR2). (b) Gene expression profiles (heat map) of the microarray analysis are shown. Genes with >1.5-fold upregulation in WT compared against control (empty vector: vec) for both CR1 and CR2 were selected and triplicate sample data (n = 3) for each gene are displayed (see Materials and Methods for more details). Gene identity is presented on the left. mRNA and probe identities of genes are presented in Figure S1. (c) Expression levels on the microarrays are shown in the bar charts.
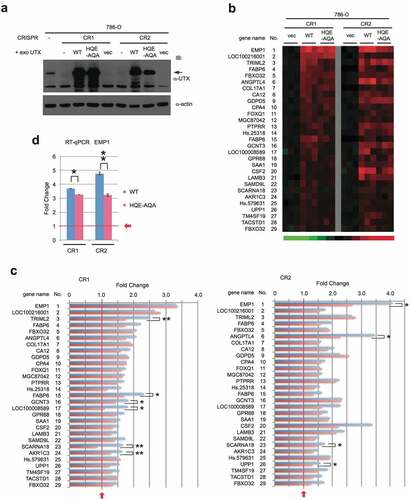
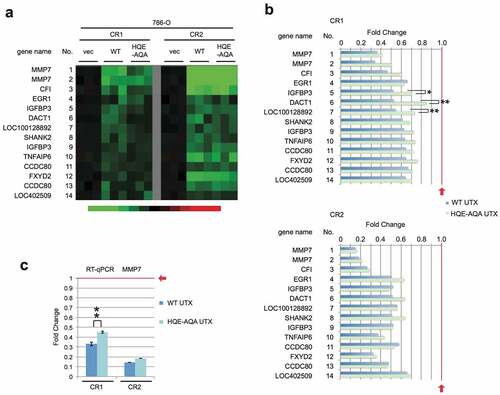
Downregulated genes (>1.5 fold down) were also identified (, S1b). All the genes downregulated by WT were also downregulated by HQE-AQA to variable extents (). Similar to the upregulated genes, the levels of downregulation were compromised for most of the genes and yet were still lower than the control vector level (=1.0). This suggests that negative regulation by UTX was also partially dependent on the catalytic activity. This was also confirmed on the correlation plots comparing WT against HQE-AQA UTX (Figure S2a,b). Expression of the most strongly downregulated gene, MMP7 (Matrix Metallopeptidase 7), was validated by RT-qPCR, which again reproduced the microarray profiling data well (). In clinical data from Morpheus, expression of MMP7 negatively correlates with that of UTX in KIRC and COAD with the correlation values of −0.12 and −0.18, respectively (Figure S3). Intriguingly, EMP1 and MMP7, which have been regarded as major oncogene candidates implicated in filtration and metastasis of cancer cells [Citation38,Citation39], are regulated in opposing manners in these cancer types. Interestingly, in a similar context, it has been shown that UTX positively regulates genes including MMP9 and MMP11 in a breast adenocarcinoma cell line MDA-MB-231 in UTX knockdown experiments [Citation23] and, conversely, negatively regulates genes including MMP9 and MMP10 in the UTX-null bladder transitional cell carcinoma cell line KU-19-19 treated with UTX mRNA-containing nanoparticles [Citation40]. The molecular basis of the negative regulation by UTX could be explained by the activation of negative transcriptional regulators or alterations of chromatin structure. As an example of the latter case, Utx loss causes alterations in H3K27 acetylation and chromatin accessibility, drives up- and down-regulation of target gene transcription, and thereby activates an ETS-mediated oncogenic programme [Citation21].
Figure 2. Expression profiles of downregulated genes in 786-O cells. (a) Similarly to , genes with >1.5-fold downregulation were selected and its heat map is shown. (b) Expression levels are shown in the bar charts with statistical values (*: p < 0.05; **: p < 0.01). (c) RT-qPCR analysis was performed for the most strongly downregulated gene MMP7.
UTX regulates a distinct set of genes in HCT116 to variable degrees of catalytic activity-dependence
We recently reported that WT UTX downregulates a set of genes associated with integrated stress response (ISR) via activation of HRI kinase [Citation34]. Here, effects of HQE-AQA UTX on expression of these genes in HCT116 were analysed. Four upregulated and thirteen downregulated genes, which were identified similarly to the experiments for 786-O, were compared between WT and HQE-AQA UTX (). Upregulation of ZBED and HAS3 was significantly attenuated for HQE-AQA (ZBED3: 31% and 24% reduction in CR1 and CR2 respectively; HAS3: 25% and 29% reduction in CR1 and CR2 respectively) (). Downregulation of DDIT3 and DDIT4 was also attenuated but weakly (DDIT4: 20% and 15% reduction in CR1 and CR2, respectively; DDIT3: 21% reduction in CR2) (), indicating that suppression of ISR via the HRI signalling pathway by UTX is mainly catalytic activity-independent. In the Morpheus dataset, correlation coefficients of expression for ISR target genes (PCK2, DDIT3, DDIT4, CHAC1, and TRIB3) with UTX in COAD are − 0.06, −0.10, −0.19, −0.25, and − 0.25, respectively (Figure S3), suggesting that these genes are also downregulated by UTX in the patient context.
Figure 3. Expression profiles of UTX-regulated genes in UTX-disrupted HCT116 cells re-expressing WT or HQE-AQA UTX. (a) Heat map is presented for up- and down-regulated genes similarly to . The WT data was previously published [Citation27], but included for side-by-side comparisons with HQE-AQA. (b) Bar charts for upregulated genes. (c) Bar charts for downregulated genes. (d) RT-qPCR analysis for representative ISR-regulated genes. n.S./c indicates not significant to empty vector ( = 1.0). *: p < 0.05; **: p < 0.01.
![Figure 3. Expression profiles of UTX-regulated genes in UTX-disrupted HCT116 cells re-expressing WT or HQE-AQA UTX. (a) Heat map is presented for up- and down-regulated genes similarly to Figure 1b. The WT data was previously published [Citation27], but included for side-by-side comparisons with HQE-AQA. (b) Bar charts for upregulated genes. (c) Bar charts for downregulated genes. (d) RT-qPCR analysis for representative ISR-regulated genes. n.S./c indicates not significant to empty vector ( = 1.0). *: p < 0.05; **: p < 0.01.](/cms/asset/54ce33cd-00a9-48b1-9a33-a378e6f94466/kepi_a_2222245_f0003_oc.jpg)
WT and HQE-AQA, but not interaction-defective mutants, suppress colony formation
In our previous report, UTX mutants interact with MLL3/4 complexes with different affinities (i.e., HQE-AQA: strong; Y375C: moderate; G137V and Δ80–397: weak or null) and G137V was unable to suppress colony formation in soft agar assays [Citation31]. In this study, we tested if UTX mutants including HQE-AQA can suppress colony formation in the same assay where they were expressed in HeLa that has undetectable levels of UTX (). In accordance with the previous result, WT efficiently diminished the number of colonies (). HQE-AQA suppressed colony formation as efficiently as WT, whereas the interaction-defective mutants (Δ80–397 and Y375C) failed to do so. Together with the previous result for the interaction-defective mutant G137V, these indicate that the interaction ability rather than the catalytic activity of UTX is critical for suppression of colony formation at least in this experimental setting. Knock-in experiments with catalytic activity-defective and MLL3/4 interaction-defective UTX mutants in an appropriate mouse tumour model will be the next steps in addressing this question.
Discussion
We show here that UTX differentially regulates gene expression in catalytic activity-dependent and -independent manners. The latter is important for the expression of a wide array of target genes and colony formation in our experimental setting, while the target specificity is highly cancer type-specific.
Numerous studies have reported UTX target genes in various normal or malignant tissue types and cell lines from several organisms. However, common targets have been rarely reported even among studies with similar experimental settings. The top-ranked UTX target genes identified for 786-O and HCT116 in this study are also different from those reported by others. We suspect that this highly context-dependent transcriptional regulation by UTX is due to its intrinsic property as a cofactor, whose specificity is mainly dependent on the transcriptional regulatory frameworks governed by target-specific DNA binding transcription factors. This context-dependent specificity could give rise to tissue- and cell type-specific regulation by UTX in development and cancer formation and complicate its role as a tumour suppressor versus oncogene issue. Consistently, the overall cancer dependency score of UTX is neutral in many cell lines including 786-O and HCT116 (0.0665 and −0.0436, respectively) according to DepMap (Broad Institute). Moreover, among various cancer types, the major target genes we identified exhibit highly divergent correlation coefficients with UTX in expression (Figure S3). Furthermore, UTX regulates the oncogene candidates EMP1 and MMP7 in 786-O cells positively and negatively, respectively ().
Evidence for the underlying mechanisms of the catalytic activity-independent action of UTX has been partially presented. Wang et al. demonstrated that UTX recruits MLL3/4 and p300 to RAR/RXR bound on target enhancers, allows proximal H3K4 monomethylation and H3K27 acetylation, and activates gene expression [Citation32]. We show here, consistent with our previous findings [Citation31], that several cancer-derived UTX substitution mutations in the TPR domain compromise interactions with MLL3/4 complexes and, in parallel, abolish the ability of UTX to suppress soft agar colony formation. In contrast, HQE-AQA, which retains the interaction ability with MLL3/4 complexes, efficiently suppresses colony formation (). Our hypothesis is that the contribution of the UTX catalytic activity to development and cancer formation is highly tissue- and cell type-specific. In certain cases, the MLL3/4 and p300 catalytic activities are functionally dominant for a wide array of genes and the critical role of UTX is to recruit MLL complexes and p300 to the target sites. UTX exerts its catalytic activity only where H3K27 demethylation is additionally required. To investigate this further, we performed analysis of the histone modification states of these dependent- and independent-gene promoter/enhancer regions using a ChIP database (ChIP-Atlas). We found that promoter/enhancer regions of the catalytic activity-dependent genes (EPM1, TRIML2, ANGPTL4, and SCARNA18; mean ratio of HQE-AQA to WT UTX = 0.67: 33% down) were heavily modified with H3K4me1, whereas those of independent genes (CPA4, FOXQ1, SAA1, and GDPD5; mean ratio of HQE-AQA to WT UTX = 1.00) were heavily modified with H3K27me3 (Figure S4a,b). These two histone modifications are signature marks of active and suppressed enhancers, respectively [Citation41]. However, in our case, both of these dependent and independent target genes were activated by UTX re-expression, suggesting that MLL3/4 complexes may be recruited by UTX also to the promoter/enhancer regions of the independent genes and mono-methylation by MLL3/4 probably has some positive effects on transcription, but demethylation by UTX is limited or dominated by the proximal presence of EZH2-containing PRC2 (Figure S5). This implies functional dominance of EZH2 over UTX. To address these mechanistic issues, knock-in experiments with catalytic activity-defective and MLL3/4 interaction-defective UTX mutants in an appropriate mouse tumour model will be the next steps.
The dominance of recruitment over catalytic activity may not be generally true for other cases of stoichiometric and, especially, non-stoichiometric interactions. For example, the histone H3K9me1/2 methyltransferase G9a and its paralog GLP form a stable stoichiometric complex, and both of these enzymes are essential for the methylation activity in vivo [Citation42,Citation43]. Catalytic activity-defective GLP mutants that retain the heterodimer formation ability with WT G9a can maintain histone methylation and gene silencing [Citation42]. A major difference from our case is that G9a and GLP have the same enzymatic activity with the same substrate specificity and both are indispensable for the enzymatic activity of the complex. One of the hallmarks of histone modification is bivalency: H3K4me3 and H3K27me3 co-occupy early development-related gene promoters or embryonic stem cells. Although Utx contribution to bivalency in development and cancer has not been conclusive [Citation44,Citation45], it is noteworthy that in maintenance or loss of bivalency, H3K27 demethylation and transcriptional regulation by UTX cannot be regarded as synonymous with each other.
The target gene- and cell-type-specific catalytic activity dependency and tumour suppressor or oncogene issues for UTX certainly impact development of anti-cancer pharmaceutical inhibitors targeting H3K27 modifications. Tazemetostat, an EZH2 inhibitor, has been approved by FDA for the treatment of metastatic or locally advanced epithelioid sarcoma and relapsed or refractory EZH2-activated follicular lymphoma (Clinical Trials, NCT01897571/NCT03456726) with a companion diagnostic test. Cancers under the control of the catalytic activity-independent target genes (i.e., heavily H3K27me3-modified genes) may be more responsive to EZH2 inhibitors. In addition, further clinical studies on INI1/SMARCB1- or BRG1/SMARCA4-deficient Rhabdoid and CNS malignant tumours are in progress (NCT02601950/NCT05407441). UTX, MLL3/4, p300, and the chromatin remodelling SWI/SNF genes (i.e., ARID1A, ARID1B, PBRM1, BRG1, INI1, etc.), all belong to the Trithorax group and function cooperatively or complementary with each other. These are frequently mutated in various cancer types (e.g., ARID1A:33% and MLL2–4:10–13% in endometrial carcinoma; UTX:26% in bladder carcinoma; PBRM1:27% in clear cell renal cell carcinoma, including nonsense, frame-shift, deletions/insertions, and splice junction mutations (Morpheus)) [Citation46]. Therefore, EZH2 inhibitors may be effective also for cancers with mutations in some of these and other Trithorax genes. Furthermore, inhibitors of H3K4 demethylases and H3K27 deacetylases that could enhance or replace the activities of MLL3/4 and p300 may also be applicable in EZH2-dependent or Trithorax-deficient cancers. Taken together, dependency on the UTX catalytic activity and the mutation status of the Trithorax genes can help predict patient response and set rational clinical trial designs.
In this study, we analysed that the catalytic activity-dependent and -independent transcriptional activation of specific target genes by UTX in our UTX knockout and re-expression system. Since other studies have observed more relevance of the independent activity to cancer formation and UTX targets are significantly heterogeneous among different experimental settings and clones, we refrain from defining the catalytic activity dependency of the selected genes and take the results of this study as one of the model cases with mechanistic implications. Further experiments in multiple settings are warranted to obtain more general understandings.
Author contributions
H.K. conceived the project. H.K., K.L.L., and L.P. designed and supervised experiments. K.L.L., H.M., and H.K. prepared the manuscript. K.L.L. conducted microarray gene expression profiling and bioinformatic analysis; W.S. performed RT-qPCR; W.S. and H.K. established UTX-disrupted cell lines and assayed for colony formation and protein expression. W.S., K.L.L., L.P., H.M., and H.K. analysed and discussed the results. All the authors except for L.P. reviewed the final manuscript.
Supplemental Material
Download Zip (1.7 MB)Acknowledgments
We thank all the lab members for technical assistance. This research is supported by the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centres of Excellence initiative. In the midst of this project, L.P. passed away to whom we dedicate this work.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The microarray gene expression profile data for 786-O and HCT116 have been deposited in the Gene Expression Omnibus (GEO, NCBI) with the accession number GSE127860 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE127860).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592294.2023.2222245
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Hong S, Cho YW, Yu LR, et al. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci, USA. 2007;104(47):18439–13. doi:10.1073/pnas.0707292104
- Agger K, Cloos PAC, Chistensen J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi:10.1038/nature06145
- Lee MG, Villa R, Trojer P, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318(5849):447–450. doi:10.1126/science.1149042
- Lan F, Bayliss PE, Rinn JL, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi:10.1038/nature06192
- Cho YW, Hong T, Hong S, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282(28):20395–20406. doi:10.1074/jbc.M701574200
- Issaeva I, Zonis Y, Rozovskaia T, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27(5):1889–1903. doi:10.1128/MCB.01506-06
- Patel SR, Kim D, Levitan I, et al. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev Cell. 2007;13(4):580–592. doi:10.1016/j.devcel.2007.09.004
- Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298:1039–1043. doi:10.1126/science.1076997
- Czemin B, Melfi R, McCabe D, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/S0092-8674(02)00975-3
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, et al. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902
- Lee S, Lee JW, Lee S-K. UTX, a histone H3-lysine 27 demethylases, acts as a critical switch to activate the cardiac developmental program. Dev Cell. 2012;22:1–13. doi: 10.1016/j.devcel.2011.11.009
- Wang C, Lee J-E, Cho Y-W, et al. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci, USA. 2012;109:15324–15329. doi:10.1073/pnas.1204166109
- Shpargel KB, Sengoku T, Yokoyama S, et al. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 2012;8(9):e1002964. doi:10.1371/journal.pgen.1002964
- van der Meulen J, Speleman F, van Vliergerghe P. The H3K27me3 demethylase UTX in normal development and disease. Epigenetics. 2014;9:658–668. doi:10.4161/epi.28298
- Wang L, Shilatifard A. UTX mutations in human cancer. Cancer Cell. 2019;35(2):168–176. doi:10.1016/j.ccell.2019.01.001.
- Nickerson ML, Dancik GM, Im KM, et al. Concurrent alterations in TERT. KDM6A, And The BRCA Pathway In Bladder Cancer. Clin Cancer Res. 2014;20:4935–4948. doi:10.1158/1078-0432.CCR-14-0330
- Ntziachristos P, Tsirigos A, Welstead GG, et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature. 2014;514(7523):513–517. doi:10.1038/nature13605
- Li X, Zhang Y, Zheng L, et al. UTX is an escape from X-inactivation tumor-suppressor in B cell lymphoma. Nat Commun. 2018;9(1):2720. doi:10.1038/s41467-018-05084-w
- Andricovich J, Perkail S, Kai Y, et al. Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell. 2018;33:512–526. doi:10.1016/j.ccell.2018.02.003
- Revia S, Seretny A, Wendler L, et al. Histone H3K27 demethylase KDM6A is an epigenetic gatekeeper of mTORC1 signalling in cancer. Gut. 2022;71:1613–1628. doi:10.1136/gutjnl-2021-325405
- Gozdecka M, Meduri E, Mazan M, et al. UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nat Genet. 2018;50(6):883–894. doi:10.1038/s41588-018-0114-z
- Shi B, Li W, Song Y, et al. UTX condensation underlies its tumor-suppressive activity. Nature. 2021;597:726–731. doi:10.1038/s41586-021-03903-7
- Kim JH, Sharma A, Dhar SS, et al. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2014;74(6):1705–1717. doi:10.1158/0008-5472.CAN-13-1896
- Xie G, Liu X, Zhang Y, et al. UTX promotes hormonally responsive breast carcinogenesis through feed-forward transcription regulation with estrogen receptor. Oncogene. 2017;36(39):5497–5511. doi:10.1038/onc.2017.157
- Wang Y, Hou N, Cheng X, et al. Ezh2 acts as a tumor suppressor in Kras-driven lung adenocarcinoma. Int J Biol Sci. 2017;13(5):652–659. doi:10.7150/ijbs.19108
- Dhar S, Gadd S, Patel P, et al. A tumor suppressor role for EZH2 in diffuse midline glioma pathogenesis. Acta Neuropathol Commun. 2022;10(1):47. doi:10.1186/s40478-022-01336-5
- Schulz WA, Lang A, Koch J, et al. The histone demethylase UTX/KDM6A in cancer: progress and puzzles. Int J Cancer. 2019;145(3):614–620. doi:10.1002/ijc.32116
- van Haaften G, Dalgliesh GL, Davies H, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi:10.1038/ng.349
- Ho AS, Kannan K, Roy DM, et al. The mutational landscape of adenoid cystic carcinoma. Nat Genet. 2013;45(7):791–798. doi:10.1038/ng.2643
- Barrows D, Feng L, Carroll TS, et al. Loss of UTX/KDM6A and the activation of FGFR3 converge to regulate differentiation gene-expression programs in bladder cancer. Proc Natl Acad Sci, USA. 2020;117(41):25732–25741. doi:10.1073/pnas.2008017117
- Kato H, Asamitsu K, Sun W, et al. Cancer-derived UTX TPR mutations G137V and D336G impair interaction with MLL3/4 complexes and affect UTX subcellular localization. Oncogene. 2020;39(16):3322–3335. doi:10.1038/s41388-020-1218-3
- Wang S-P, Tang Z, Chen C-W, et al. A UTX-MLL4-p300 transcriptional regulatory network coordinately shapes active enhancer landscapes for eliciting transcription. Mol Cell. 2017;67:308–321. doi:10.1016/j.molcel.2017.06.028
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi:10.1038/nmeth.3047.
- Kitajima S, Sun W, Lee KL, et al. A KDM6 inhibitor potently induces ATF4 and its target gene expression through HRI activation and partly by UTX inhibition. Sci Rep. 2021;11:4538. doi:10.1038/s41598-021-83857-y
- Sun W, Kato H, Kitajima S, et al. Interaction between von Hippel-Lindau protein and fatty acid synthase modulates hypoxia target gene expression. Sci Rep. 2017;7(1):7190. doi:10.1038/s41598-017-05685-3
- Oki S, Ohta T, Shioi G, et al. ChiP-Atlas: a data-mining suite powered by full integration of public ChiP-seq data. EMBO Rep. 2018;19:e46255. doi:10.15252/embr.201846255
- Dalgliesh GL, Furge K, Greenman C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463(7279):360–363. doi:10.1038/nature08672
- Ahmat Amin MKB, Shimizu A, Zankov DP, et al. Epithelial membrane protein 1 promotes tumor metastasis by enhancing cell migration via copine-III and Rac1. Oncogene. 2018;37(40):5416–5434. doi:10.1038/s41388-018-0286-0
- Liao H-Y, Da C-M, Liao B, et al. Roles of matrix metalloproteinase-7 (MMP-7) in cancer. Clin Biochem. 2021;92:9–18. doi:10.1016/j.clinbiochem.2021.03.003
- Kong N, Zhang R, Wu G, et al. Intravesical delivery of KDM6A-mRNA via mucoadhesive nanoparticles inhibits the metastasis of bladder cancer. Proc Natl Acad Sci, USA. 2022;119(7):e2112696119. doi:10.1073/pnas.2112696119
- Herz H-M, Mohan M, Garruss AS, et al. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes&dev. 2012;26:2604–2620. doi:10.1101/gad.201327.112
- Tachibana M, Matsumura Y, Fukuda M, et al. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. Embo J. 2008;27(20):2681–2690. doi:10.1038/emboj.2008.192
- Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes&dev. 2011;25:781–788. doi: 10.1101/gad.2027411
- Dhar SS, Lee S-H, Chen K, et al. An essential role for UTX in resolution and activation of bivalent promoters. Nucleic Acids Res. 2016;44(8):3659–3674. doi:10.1093/nar/gkv1516
- Shpargel KB, Starmer J, Yee D, et al. KDM6 demethylase independent loss of histone H3 lysine 27 trimethylation during early embryonic development. PLoS Genet. 2014;10:e1004507. doi: 10.1371/journal.pgen.1004507
- Baylin SB, Jones PA. Epigenetic determinant of cancer. Cold Spring Harb Perspect Biol. 2016;8:a019505. doi: 10.1101/cshperspect.a019505