ABSTRACT
Osteosarcoma, originating from primitive bone-forming mesenchymal cells, is the most common malignant bone tumour among children and adolescents. N6-methyladenosine (m6A), the most ubiquitous type of posttranscriptional modification, is a methylation that occurs in the N6-position of adenosine. m6A dramatically affects the splicing, export, translation, and stability of various RNAs, including mRNA and noncoding RNAs (ncRNAs). Increasing evidence suggests that ncRNAs, especially microRNAs (miRNA), long noncoding RNAs (lncRNA), and circular RNAs (circRNAs), regulate the m6A modification process by affecting the expression of m6A-associated enzymes. m6A modification interactions with ncRNAs provide new perspectives for exploring the underlying mechanisms of tumorigenesis and progression. In the current review, we summarized the expression and biological functions of m6A regulators in osteosarcoma. At the same time, the present review systematically elucidated the functional and mechanical interactions between m6A modification and ncRNAs in osteosarcoma. In addition, we discussed the effect of m6A and ncRNAs in the tumour microenvironment and potential clinical applications of osteosarcoma.
Introduction
Osteosarcoma, originating from primitive bone-forming mesenchymal cells, is the most common malignant bone tumour in children and adolescents and usually occurs in the epiphysis of the long bones; therefore, osteosarcoma is characterized by rapid growth and fast progression [Citation1]. There is growing evidence that osteosarcoma may be associated with cancer stem cells (CSCs), DNA repair-related gene defects, tumour inhibition pathways, and gene changes [Citation2]. Early diagnosis of osteosarcoma remains difficult, and it is compelling to further explore the molecular mechanism of osteosarcoma and to discover new therapeutic targets to better guide the treatment of osteosarcoma.
N6-Methyladenosine (m6A) is methylation that occurs in the N6-position of adenosine, which is one of the most ubiquitous types of posttranscriptional modification [Citation3]. More than 12,000 m6A sites have been found in more than 7000 human genes, which are rich in RRACH (R=G or A, H=A, C or U) shared sequences. These sequences usually exist in the termination codon and 3´ untranslated region (3´UTR) [Citation4,Citation5]. In addition, m6A modification sites can also exist in unique motifs of different noncoding RNAs (ncRNAs), including long noncoding RNAs (lncRNAs), circular RNAs (circRNAs), and microRNAs (miRNAs) [Citation6]. As a dynamic and reversible modification, m6A modification is mainly regulated by three types of proteins: m6A methyltransferases (m6A writers), m6A demethylases (m6A erasers), and m6A-binding proteins (m6A readers). ‘Writers,’ also called the m6A methyltransferase complex (MTC), catalyse the modification of m6A. Demethylases, alternatively named ‘erasers,’ are capable of removing methylation in DNA and histones. Potential m6A selective binding proteins, also named ‘readers,’ are in charge of recognizing methylated RNAs to perform biological functions ().
Figure 1. Dynamic regulation of RNA m6A levels by m6A and the known functions of m6A in the regulation of RNA metabolism. m6A modifications are catalysed by the methyltransferase complex consisting of METTL3 and METTL14, as well as their cofactors WTAP, KIAA1429 and RBM15/15B (writers). The removal of m6A modifications relies on the demethylases FTO and ALKBH5 (erasers). m6A modifications are functionally facilitated by the m6A binding proteins YTHDF1–3, YTHDC1–2, IGF2BP1–3, ELAV1, and HNRNPs (readers).
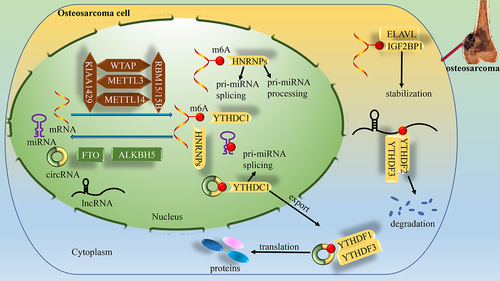
With the progress of sequencing technology, a large number of ncRNAs, such as miRNAs, circRNAs, and lncRNAs, have been found [Citation7]. ncRNAs, with limited ability to encode proteins or peptides, demonstrate broad potential in gene regulation and tumour biology [Citation8]. There are many m6A modification sites in ncRNAs (including circRNAs, lncRNAs, and miRNAs), and m6A can highly modify ncRNAs [Citation9]. m6A not only regulates the cleavage, localization, trafficking, stability and degradation of ncRNAs [Citation3,Citation10,Citation11], but also interacts with ncRNAs and affects the biological functions of cells, including the proliferation, infiltration and metastasis of tumour cells; apoptosis; and chemoresistance [Citation12,Citation13]. Interestingly, ncRNAs can also target m6A-associated proteins during tumorigenesis and progression, regulate the interaction between m6A related proteins and downstream target mRNA transcripts, and control the degradation, translation and expression of target mRNAs [Citation14,Citation15]. Discussion of the role of m6A-ncRNA interactions in osteosarcoma could provide new directions for the diagnosis and treatment of osteosarcoma.
In the current review, we provide an update on the aberrant expression and function of m6A-related enzymes in osteosarcoma. Additional special emphasis has also been placed on the interaction of the m6A modification with regulatory ncRNAs in osteosarcoma, which includes the m6A modification on regulatory ncRNAs and the regulation of m6A modification by ncRNAs.
Aberrant m6A regulation in osteosarcoma
In osteosarcoma, the global abundance of m6A and the expression levels of its regulators (including writers, erasers and readers) are often dysregulated.
Aberrant global m6A abundance in osteosarcoma
Global m6A levels in osteosarcoma are increased during the development of osteosarcoma, and this imbalance may be associated with osteosarcoma development and clinical outcomes.
Quantitative analysis of m6A RNA methylation by liquid chromatography tandem mass spectrometry (LC‒MS/MS) revealed significantly increased levels of m6A RNA modification in osteosarcoma tissues. The level of m6A modification in osteosarcoma tissues from patients with lung metastases was significantly higher than that in patients without lung metastases [Citation16]. Suggesting that m6A modification is closely related to poor prognosis. Similarly, at the cellular level, Yuan et al. demonstrated that osteosarcoma cells have higher m6A levels than normal osteoblasts using m6A ELISA and immunofluorescence (IF) [Citation17].
Aberrant expression of m6A regulators in osteosarcoma
The dynamic reversible process of m6A is mainly related to three types of m6A regulators: ‘writers,’ ‘erasers’ and ‘readers.’ ‘writers,’ mainly composed of methyltransferase-like 3, 14 and 16 (METTL3, METTL14 and METTL16) and their cofactors, Wilms tumour 1-associated protein (WTAP), RNA binding motif protein 15 (RBM15), and Vir-like m6A methyltransferase associated (KIAA1429) [Citation18–21]. Fat mass and obesity-associated protein (FTO) and alkylation repair homolog protein 5 (ALKBH5) are the two m6A demethylases that have been identified so far [Citation22,Citation23]. Current m6A readers include the YT521-B homology (YTH) domain family (YTHDF1, YTHDF2, and YTHDF3), YTH domain-containing proteins (YTHDC1 and YTHDC2), heterogeneous nuclear ribonucleoprotein (HNRNP) protein families, eukaryotic translation initiation factor 3 (eIF3), Staphylococcal nuclease domain-containing protein 1 (SND1), embryonic lethal abnormal vision Drosophila-like 1 (ELAVL1), and insulin-like growth factor-2 mRNA-binding proteins (IGF2BP1, IGF2BP2, and IGF2BP3) [Citation24–27]. Aberrant m6A regulators closely participate in the tumorigenesis and development of osteosarcoma.
To identify the functional role of m6A regulators in osteosarcoma, genome-wide gene expression profiling was recently performed among mesenchymal stem cells, osteosarcoma cells, and osteosarcoma cell lines. It was revealed that m6A ‘writers’ such as METTL3 and RBM15, ‘readers’ such as YTHDF1, YTHDF2, YTHDC1, and HNRNP2B1, and ‘erasers’ such as FTO were upregulated in osteosarcoma. Upregulated m6A regulators in osteosarcoma are strongly associated with poor survival and can be used as diagnostic biomarkers and potential targeted therapeutic targets in osteosarcoma [Citation28].
Additionally, the m6A regulators that are implicated in osteosarcoma metastasis were further explored. The expression levels of mRNAs from osteosarcoma tumour tissues with and without metastasis were downloaded from TCGA and reanalysed by an Illumina human-6 v2.0 expression beadchip. Significant differential expression of METTL14, VIRMA, METTL3, WTAP and YTHDC1 was found in osteosarcoma samples with metastasis compared with those in osteosarcoma samples without metastasis, suggesting that m6A RNA regulators are involved in the metastasis of osteosarcoma [Citation29]. Targeting these m6A regulatory proteins may inhibit the migration and metastasis of osteosarcoma.
MG63/DXR refers to the enrichment of a multidrug-resistant cell population by gradually increasing the concentration of doxorubicin in the osteosarcoma cell line MG63. In studies of the expression of m6A methylation-related enzymes in MG63/DXR cells, it was found that METTL3 and ALKBH5 were upregulated at both the mRNA and protein levels, with no significant change in FTO, and METTL14 was upregulated at the protein level, suggesting that m6A regulators play a regulatory role in chemoresistance [Citation30]. Targeting m6A modification mediated by METTL3, METTL14 and ALKBH5 May be a promising adjuvant treatment strategy for patients with chemically resistant osteosarcoma.
In conclusion, in osteosarcoma cell lines and tissues, the global abundance of m6A and the expression level of its regulatory factors are often dysregulated, and dysregulated regulators were strongly associated with poor survival in osteosarcoma and mediated invasive metastasis and chemoresistance in osteosarcoma, which may provide a promising diagnostic biomarker and potential targeted therapeutic strategy for patients with osteosarcoma. Regarding the cause of aberrant m6A regulation, it may be due to base mutations on mRNAs that result in the addition or loss of m6A sites, or environmental factors that can reprogram the episodic transcriptome, or dysregulation of the m6A machinery leading to upregulation of oncogenes or downregulation of tumour suppressor genes [Citation31].
Biological functions of m6A regulators in osteosarcoma
Normal biological processes and development depend on maintaining appropriate levels of m6A modification on RNA, and malfunction of m6A regulators is often associated with cancer [Citation32]. Here, we summarize the different regulators of m6A to exert biological processes in osteosarcoma () ().
Figure 2. METTL3 regulates osteosarcoma biology by acting on TRAF6, ATAD2 and LEF1. METTL14 regulates osteosarcoma biological processes by acting on MN1 and caspase-3. WTAP regulates osteosarcoma biological processes through HMBOX1. METTL3, METTL14 and YTHDF regulate osteosarcoma biological processes through TRIM7. FTO regulates osteosarcoma biological processes through KLF3 and DACT1. ALKBH5 regulates the biological process of osteosarcoma through SOCS3 and USP22/RNF40. YTHDF1 regulates the biological process of osteosarcoma through CONT7. ELAVL1 regulates the biological process of osteosarcoma through DRG1.
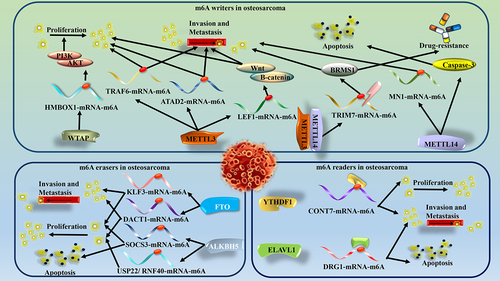
Table 1. Roles of m6A regulators in osteosarcoma.
Biological functions of m6A writers in osteosarcoma
METTL3 was the first identified m6A methyltransferase [Citation50]. A recent study showed that METTL3 promoted the progression of osteosarcoma cells by regulating the m6A level of lymphoid enhancer binding factor 1 (LEF1) and activating the Wnt/β-catenin signalling pathway [Citation36]. Similarly, knockdown of METTL3 suppressed the expression of ATPase family AAA domain containing 2 (ATAD2), thereby inhibiting the proliferation and invasion ability of osteosarcoma cells [Citation33]. In another study, METTL3 positively regulated TNF receptor-associated factor 6 (TRAF6) expression and promoted TRAF6-mediated metastasis of osteosarcoma. TRAF6 also reversed the inhibitory effect of sh-METTL3 on the invasion, metastasis and EMT of osteosarcoma cells [Citation35]. METTL3 has also been reported to be involved in metabolic processes in osteosarcoma. Ubiquitin-specific proteases 13 (USP13) regulates glycolytic reprogramming and proliferation in osteosarcoma by deubiquitinating METTL3 at K488. Subsequently METTL3 increases autophagy-related gene 5(ATG5) mRNA stabilization in an m6A-modified manner, thereby promoting oncogenic autophagy [Citation39]. In addition to increasing the stability of mRNA, METTL3 can also increase the stability of lncRNA and promote the malignant progression of osteosarcoma. METTL3 was found to mediate the expression of differentiation antagonizing nonprotein coding RNA (DANCR) in osteosarcoma, which in turn promoted the proliferation, invasion and metastasis of osteosarcoma cells [Citation51]. As reported above, METTL3 as an oncogenic gene mediates the proliferation, invasion, and metastasis in osteosarcoma.
As the most important catalytic subunit, the catalytic activity of METTL3 is strongly dependent on METTL14 [Citation52]. Liu and associates demonstrated that METTL14 was expressed at low levels in osteosarcoma. Inhibition of METTL14 significantly promoted proliferation, migration, and invasion while promoting apoptosis in osteosarcoma cells via regulation of the caspase-3 pathway [Citation41]. Controversially, however, METTL14 was demonstrated to be highly expressed in osteosarcoma and associated with poor prognosis. METTL14 mediates the stability and translation of MN1 mRNA, thereby promoting the proliferation and metastasis of osteosarcoma cells [Citation16]. Some emerging evidence has also shown that METTL14 works together with METTL3 to reduce the expression of DIRAS1 by regulating the ERK pathway and enhancing proliferation, invasion, and migration while blocking apoptosis in osteosarcoma [Citation53].
WTAP, which lacks methylation activity, always interacts with the METTL3-METTL14 complex and affects the deposition of m6A in cells [Citation18]. The absence of WTAP reduces the binding ability of METTL3 to RNAs, indicating that WTAP is essential for the recruitment of the m6A methyltransferase complex [Citation3]. As a splicing factor in mammals, WTAP is reported to be highly expressed in osteosarcoma, and WTAP is related to the poor prognosis of osteosarcoma patients. Mechanistically, WTAP targets and regulates the 3´UTR of homeobox containing 1 (HMBOX1), and then HMBOX1 activates the PI3K/Akt signalling pathway to regulate the proliferation and metastasis of osteosarcoma [Citation40]. For ncRNAs, WTAP is capable of modifying the methylation of the lncRNA FOXD2 adjacent opposite strand RNA 1 (FOXD2-AS1), which accelerates the proliferation, migration and metastasis of osteosarcoma cells in vitro and in vivo [Citation54].
RBM15 is a member of the SPEN (split-end) protein family and interacts with RNA by binding to spliceosomal components [Citation55]. A recent study reported that circ-CTNNB1 and RBM15 interact through the RRM1 structural domain to mediate the m6A modification of hexokinase 2 (HK2), glucose-6-phosphate isomerase (GPI) and phosphoglycerate kinase 1 (PGK1) and promote the aerobic glycolytic process in osteosarcoma cells [Citation56]. The interaction between circ-CTNNB1 and RBM15 relies on the RRM1 domain, and targeting the RRM1 domain provides a new direction for the treatment of osteosarcoma.
Recent research has identified METTL16 as a crucial m6A methyltransferase that plays a significant role in embryonic development and maintaining S-adenosylmethionine (SAM) balance [Citation57]. According to Cheng et al., METTL16 is highly expressed in osteosarcoma. It is responsible for promoting tumour growth and metastasis by facilitating the degradation of vacuolar protein sorting protein 33b (VPS33B) mRNA [Citation42]. Limited studies have been conducted on the role of METTL16 in cancer, and this study provides a new direction for treating osteosarcoma.
KIAA1429, a scaffold connecting the catalytic core components METTL3 and METTL14, is also up-regulated in osteosarcoma [Citation58]. KIAA1429 promotes osteosarcoma proliferation, invasion, and migration through the JAK2/STAT3 signalling pathway [Citation43]. However, whether the interaction between KIAA1429 and the JAK2/STAT3 signalling pathway is in an m6A-modified manner has yet to be determined and requires more in-depth studies.
Biological functions of m6A “erasers” in osteosarcoma
FTO, the first identified m6A demethylase, located on chromosome 16, can catalyse the demethylation of m6A in a ferrous iron-dependent manner [Citation22]. It has been reported that FTO has higher expression in osteosarcoma tissue, where it can mediate the m6A demethylation of Dapper1/Dpr1 (DACT1), thereby reducing the RNA stability and protein expression of DACT1 and promoting the growth and metastasis of osteosarcoma through the DACT1/Wnt signalling axis [Citation44]. In another study, FTO regulated Krüppel-like factor 3 (KLF3) expression in an m6A-dependent manner, reduced the stability of KLF3 mRNA, promoted the proliferation and invasion of osteosarcoma cells, and inhibited their apoptosis [Citation45].
ALKBH5 is an m6A-specific demethylase that can directly abrogate m6A modification of adenosine [Citation23]. In one study, the protumorigenic function of ALKBH5 was mediated by regulating m6A of histone deubiquitination ubiquitin-specific peptidase 22 (USP22) and ubiquitin ligase RING finger protein 40 (RNF40). Silencing ALKBH5 makes the mRNA of USP22 and RNF40 unstable, thus leading to decreased gene expression related to the cell cycle, replication and DNA damage repair in osteosarcoma cells [Citation46]. ALKBH5 upregulated the expression of plasmacytoma variant translocation 1 (PVT1) via inhibition of its degradation, therefore promoting proliferation and tumour growth in osteosarcoma [Citation59]. Controversially, another study claimed that ALKBH5, acting as a tumour suppressor in osteosarcoma, was downregulated and inversely controlled cell growth, migration and invasion, triggering cell apoptosis by altering the m6A methylation levels of pre-miR-181b-1 and Yes1-associated transcriptional regulator (YAP) mRNA [Citation17]. In addition, ALKBH5 can also increase suppressor of cytokine signalling-3 (SOCS3) expression through an m6A-YTHDF2-dependent mechanism to inactivate the signal transducer and activator of transcription 3 (STAT3) pathway, which in turn inhibits cell proliferation and the cell cycle and promotes apoptosis in osteosarcoma [Citation47].
Biological functions of m6A “readers” in osteosarcoma
YTH family proteins are important molecules in m6A related epigenetic cancers. Various studies have shown that different YTH proteins play different roles in tumour development. However, there is still controversy over whether it is an oncogene or a tumour suppressor gene [Citation60]. In a recent study, it was demonstrated that YTHDF1 was upregulated in osteosarcoma tissues and cell lines. YTHDF1 promotes the translation of CCR4-NOT transcription complex subunit 7 m(CONT7) in an m6A-dependent manner and can promote the proliferation, migration and invasion abilities of osteosarcoma cells through the METTL3-CONT7-YTHDF1 axis [Citation37]. Previous studies have found that YTHDF2 can mediate RNA degradation, which has been confirmed in osteosarcoma [Citation38]. Knockdown of YTHDF2 increase the mRNA level of tripartite motif containing 7 (TRIM7) in osteosarcoma cells, prolong the half-life of TRIM7 mRNA in HOS and MG63 osteosarcoma cells, and then mediate the invasion and metastasis of osteosarcoma [Citation38]. YTHDF3, is also highly expressed in osteosarcoma. YTHDF3 enhances the stability of PTEN directly interacts with phosphoglycerate kinase 1 (PGK1) mRNA in an m6A-dependent manner, promoting the proliferation and aerobic glycolysis of osteosarcoma cells [Citation48]. Similarly, YTHDC1 expression levels are increased in osteosarcoma. Overexpression of YTHDC1 reversed the inhibition of RNA and protein levels of 3-phosphoinositide-dependent protein kinase 1 (PDPK1) by miR-451a, which in turn promoted the proliferation, invasion, metastasis and EMT of osteosarcoma [Citation61]. Overall, targeting the YTH domain is a new direction for targeted treatment of osteosarcoma.
IGF2BPs have been reported to enhance mRNA stability and mediate translation in an m6A dependent manner [Citation27]. In osteosarcoma, IGF2BP1 binds to the oestrogen-related receptor alpha (ERRα) 3´-UTR in an m6A manner, enhancing mRNA stability, which in turn leads to ATP production, glucose depletion, lactate production, and Doxorubicin (Dox) resistance in osteosarcoma cells [Citation49]. Targeting glucose metabolism through the IGF2BP1/ERRα axis could be a potential therapeutic strategy for overcoming Dox resistance in osteosarcoma.
ELAVL1 is one of the most abundant mRNA binding proteins in eukaryotic cells, and ELAVL1 can bind to the AU-rich elements in mRNA [Citation62]. Developmentally regulated GTP binding protein 1 (DRG1), a tumour initiator, increases cell viability while inhibiting apoptosis in osteosarcoma. ELAVL1, which works cooperatively with METTL3, increases the stability of DRG1 mRNA via m6A modification and indirectly upregulates the protein level of DRG1 [Citation34].
SND1 is a newly discovered m6A reader. Recent studies have shown that SND1 can bind to circ0024831. Overexpression of SND1 reverses the negative regulatory effect of circ0024831 on osteosarcoma and promotes the progression of osteosarcoma [Citation63].
In conclusion, m6A regulators can act as both oncogenes and tumour suppressors in osteosarcoma. Both increased and decreased levels of modified m6A may promote proliferation, invasion and metastasis, apoptosis and chemoresistance in osteosarcoma, the exact mechanism of which is unclear and may be related to different targets or signalling pathways.
Mutual regulation between m6A and ncRNA in osteosarcoma
An increasing number of studies have reported that m6A interacts with ncRNAs that are closely related to the occurrence and development of osteosarcoma () (), and the results are as follows.
Figure 3. Mutual regulation between m6A and ncRNA in osteosarcoma. (a) ALKBH5 weakens the m6A methylation modification of pre-miR-181b-1 and enhances the expression levels of both pre-miR-181b-1 and miR-181-5p. YTHDF2 recognizes pre-miR-181b-1 and enhances its stability, and YTHDF1 recognizes downstream YAP and promotes its translation, together regulating osteosarcoma progression. miR-451a regulates osteosarcoma progression by activating the AKT/mTOR pathway through regulation of YTHDC1-mediated m6A methylation. (b) pre-mRNA forms circRNA by back splicing. circ_0001105 regulation of osteosarcoma progression by sponging miR-766 and activating YTHDF2 expression. METTL3 enhances circNRIP1 expression through m6A modification, and circNRIP1 regulates osteosarcoma progression by sponging miR-199a to promote FOXC2 expression. circ0024831 directly binds to the tudor structural domain of SND1, blocking the recognition of m6A-modified RNA by SND1 and regulating osteosarcoma progression. circ-CTnNB1 interacts with RBM15 and subsequently promotes the expression of HK2, GPI and PGK1 through m6A modification to regulate osteosarcoma progression. (c) ALKBH5 inhibits the degradation of PVT1 and inhibits the binding of YTHDF2 to PVT1, regulating osteosarcoma progression. WTAP increases the stability of FOXD2-AS1 and regulates osteosarcoma progression. METTL3 increases the stability of DANCR and regulates osteosarcoma progression.
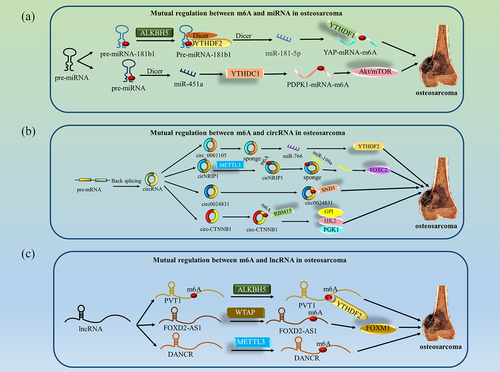
Table 2. Mutual regulation between m6A and ncRNA in osteosarcoma.
Mutual regulation between m6A and miRNA in osteosarcoma
miRNAs are small and abundant ncRNAs, approximately 20 nucleotides in length, involved in gene silencing or posttranscriptional gene expression regulation [Citation66]. During primary miRNA processing, DiGeorge syndrome critical region 8 (DGCR8) is essential to initiate miRNA maturation. m6A modification promotes miRNA biosynthesis mainly by affecting the integration between DGCR8 and pri-miRNA [Citation67]. miRNAs also have the ability to affect m6A levels involved in diverse biological processes. Modulation of the expression of the miRNA biogenesis enzyme Dicer or miRNA can alter the m6A abundance on their target transcripts [Citation68].
In osteosarcoma, m6A promotes pre-miRNA processing and maturation. A recent study found that pre-miR-181b-1 could be methylated by ALKBH5, which in turn affected its maturation in the cytoplasm. Overexpression of ALKBH5 decreased the expression of pre-miR-181b-1 and miR-181b-5p in osteosarcoma cells. ALKBH5 acts as a tumour suppressor and inhibits the proliferation, invasion and metastasis of osteosarcoma through the pre-miR-181b-1/miR-181b-5p/YAP axis [Citation17].
In addition, miRNAs modulate the m6A abundance on their target transcripts in osteosarcoma. In the latest research report, miR-451a was found to exert an inhibitory effect in osteosarcoma. Overexpression of miR-451a was found to significantly reduce the expression of YTHDC1. YTHDC1 regulates the methylation of 3-phosphoinositide-dependent protein kinase 1 (PDPK1), a downstream target gene of miR-451a. miR-451a-mediated YTHDC1 regulates the stability of PDPK1 mRNA through m6A dependence, thereby inhibiting the proliferation, invasion, metastasis and EMT of osteosarcoma [Citation61].
In summary, m6A-miRNA interactions have been found to regulate osteosarcoma progression. m6A modifications affect osteosarcoma by influencing miRNA biosynthesis. miRNAs can also regulate the abundance of m6A on their target transcripts to influence target genes, thereby regulating osteosarcoma progression.
Mutual regulation between m6A and circRNA in osteosarcoma
circRNAs are novel endogenous ncRNAs that are usually generated by pre-mRNA back-splicing, so they are circRNAs that lack 3´ and 5´ ends [Citation69]. Emerging evidence suggests that m6A modification promotes the cytoplasmic export, translation, and degradation of circRNAs [Citation70–72]. In addition, dysregulation of circRNAs also affects m6A levels [Citation73]. Moreover, circRNAs act as miRNA sponges to competitively bind with miRNAs and affect their activities and the expression of their downstream target genes, which in turn indirectly regulate m6A modification [Citation74].
m6A modification of circRNAs is crucial in the occurrence and development of osteosarcoma. Meng concentrated on the functional role of circNRIP1 in osteosarcoma. They found that circNRIP1 was upregulated in osteosarcoma tissues. Knockdown of circNRIP1 inhibited proliferation and metastasis while promoting apoptosis in osteosarcoma cells. They also revealed that METTL3 could elevate the expression level of circNRIP1 through m6A modification. Further analysis revealed that METTL3-induced circNRIP1 exerted an oncogenic role in osteosarcoma by sponging miR-199a [Citation65].
In addition, circRNAs regulate m6A modification. A recent study revealed that the nanomaterial circ0024831 could directly bind to the Tudor domain of SND1 in the cytoplast to block the recognition of m6A-modified RNA by SND1 and inhibit the occurrence and development of osteosarcoma [Citation63]. circ-CTNNB1 is highly expressed in osteosarcoma. circ-CTNNB1 regulates the m6A modification of aerobic glycolytic genes through direct binding to RBM15, leading to more stable mRNA and activation of target genes [Citation56]. Simultaneous, circ_0001105 overexpression can significantly reduce the migration and invasion ability of osteosarcoma cells. Further analysis found that circ_0001105 acted as a sponge of miR-766 to alleviate the inhibitory effect of miR-766 on its target YTHDF2 and thus regulate the progression of osteosarcoma [Citation64].
In conclusion, m6A modification can regulate the development of osteosarcoma through circRNA – miRNA–mRNA, and circRNAs act as miRNA sponges that can also affect m6A regulatory proteins. Additionally, circRNAs can bind directly to m6A-related proteins, which in turn can affect the m6A levels of their target genes.
Mutual regulation between m6A and lncRNA in osteosarcoma
lncRNAs generally refer to transcripts with a length of more than 200 nucleotides that cannot encode proteins [Citation66]. lncRNAs, interacting with other molecules, participate in histone modification, regulation of gene transcription and translation, RNA stability, RNA splicing and other processes [Citation75]. m6A methylation can act as an RNA structural switch, modulating the structure of RNA to affect RNA‒protein interactions [Citation10]. m6A methylation is also able to participate in the lncRNA-mediated ceRNA model to regulate the activity and biological function of miRNAs [Citation76]. In addition, m6A methylation can promote X inactivation-specific transcript (XIST)-mediated gene silencing [Citation77]. Previous research has shown that lncRNAs interact with m6A to participate in the progression of osteosarcoma.
lncRNA PVT1 is a functional lncRNA with carcinogenic effects [Citation78]. It was reported that ALKBH5 was upregulated in osteosarcoma tissues and cells. ALKBH5 upregulated the expression of PVT1 via inhibition of its degradation, therefore promoting proliferation and tumour growth in osteosarcoma [Citation59]. Another study showed that lncRNA FOXD2 adjacent opposite strand RNA 1 (FOXD2-AS1) accelerated osteosarcoma cell migration, proliferation and tumour growth in vitro and in vivo. An m6A modification site was identified on the 3´UTR of FOXD2-AS1, and WTAP was able to act on this site to promote methylation modification, which in turn enhanced the stability of FOXD2-AS1 transcripts and subsequently regulated downstream target Forkhead box M1 (FOXM1) mRNA and promoted osteosarcoma progression [Citation54]. In addition, there is also evidence that lncRNA DANCR is a potential target of METTL3. Silencing METTL3 can inhibit the expression of DANCR in osteosarcoma. Further analysis showed that METTL3 mediates the stability of DANCR and then regulates the proliferation, invasion and metastasis of osteosarcoma cells [Citation51].
In general, the regulatory mechanism between m6A and lncRNAs in osteosarcoma is that m6A modification regulates the stability and degradation of lncRNAs.
m6A and ncRNA in the osteosarcoma microenvironment
The tumour microenvironment (TME), which is governed by the intrinsic mechanisms of tumorigenesis and epigenetic modification, has become a research hotspot in recent years. The TME can be described by hypoxia, metabolic dysregulation, immune escape and chronic inflammation [Citation79]. m6A with ncRNAs playing a role in the osteosarcoma TME has also been reported.
m6A with ncRNAs can regulate the osteosarcoma glycolytic process, causing metabolic dysregulation of tumour microenvironment. In osteosarcoma, circ-CTNNB1 interacted with RBM15 through the RRM1 domain and increased the m6A levels of the target genes GPI, hK2 and PGK1, which in turn drove the osteosarcoma glycolytic process by increasing glucose uptake, lactate production and ATP levels in 143B and MG-63 cells [Citation56].
m6A and ncRNA participate in tumour immunity in osteosarcoma. In one study, 88 osteosarcoma samples were extracted from the TCGA database and divided into two subgroups: metastatic (22 specimens) and nonmetastatic (65 specimens). A total of seven m6A-associated lncRNAs (including TNS1-AS1, WWC2-AS1, TFPI2-DT, LINC01474, LINC00910, LINC01982 and LINC00538) were screened and were closely associated with the prognosis of osteosarcoma patients. TNS1-AS1 and TFPI2-DT were found to be positively correlated with memory B cells and naïve B cells. LINC01474 had a positive correlation with CD8 T cells; however, LINC00910 was negatively related to CD8 T cells. Moreover, LINC00538 was positively correlated with resting dendritic cells and negatively linked to activated dendritic cells in our results. m6A-associated lncRNAs are closely associated with the immune microenvironment of osteosarcoma tumours and may influence tumour occurrence and progression [Citation80]. These findings reveal new directions in the role of m6A and lncRNA in the immune microenvironment of osteosarcoma and demonstrate that m6A modification may be a potential therapeutic target for anti-tumour immunotherapy.
Tumour microenvironment is still the focus of cancer research. Many studies have demonstrated that ncRNAs are key regulators of the osteosarcoma microenvironment. For example, miR-21 can regulate the tumour microenvironment in osteosarcoma by targeting specific molecules in tumour cells, endothelial cells [Citation81]. However, only a few studies have focused on the function of m6A modified ncRNA in tumour microenvironment. It is possible that single-cell sequencing technology is used to scrutinize the m6A-modified ncRNAs in osteosarcoma microenvironment.
Clinical implications of m6A and lncRNA in osteosarcoma
Numerous studies have shown that m6A regulatory factors are closely associated with clinical features and chemotherapy resistance in osteosarcoma. Dysregulated m6A regulators may serve as prognostic markers and potential therapeutic targets for osteosarcoma. Recent studies have found that RBM15 is highly expressed in osteosarcoma, while the expression level of RBM15 is negatively correlated with the drug sensitivity of Denileukin Diftitox Ontak [Citation82]. It is possible to determine the sensitivity of osteosarcoma patients to Denileukin Diftitox Ontak by testing the expression level of RBM15. FTO is upregulated in osteosarcoma tissues, and targeting FTO inhibits osteosarcoma growth and metastasis. Entacapone, a traditional drug used for treating Parkinson’s disease, can inhibit the malignant progression of osteosarcoma by inhibiting the FTO/DACT1 axis [Citation44,Citation83]. Therefore, FTO is a potential biomarker for osteosarcoma, and the FTO inhibitor entacapone has the potential clinical use as a therapeutic agent for osteosarcoma. In another study, the knockdown of YTHDF2 increased the expression of TRIM7 and increased the resistance of osteosarcoma to adriamycin and methotrexate treatment [Citation38]. DNA damage repair proteins are essential players in radio-chemotherapy resistance, and the expression of these proteins is closely related to the m6A modification [Citation84,Citation85]. METTL3 plays an essential role in the UV-induced DNA damage response in osteosarcoma cells. The knockdown of METTL3 results in delayed repair of UV-induced cyclobutene pyrimidine dimers and increased sensitivity to radiation [Citation86]. Abnormal expression of m6A regulatory factors in patients’ tissues, cells, plasma and exosomes, may serve as biomarkers indicative of osteosarcoma cellular characteristics, providing an early and non-invasive method for the detection of osteosarcoma and offering new insights into useful targets for the diagnosis, treatment and prognosis of osteosarcoma.
The interaction between m6A modification and ncRNA may be a breakthrough for targeted osteosarcoma treatment. m6A modification tends to occur in a subset of RRACH motifs, and targeting the shared sequence, RRACH may block the binding of m6A to ncRNAs [Citation14]. With the development of CRISPR technology, many editing systems targeting m6A have emerged. In CRISPR/Cas9 m6A editing systems, fusion proteins of METTL3 and METT14 catalytic structural domains (M3-M14) may be tagged to the N-terminus of RNAs targeting dCas9 mutants, and the dCas9-M3-M14 complex is targeted to specific RNA sequences by sgRNA and PAM antisense oligomers (PAMers). In addition, RNA targeting dCas9 May be fused to the m6A demethylase FTO or ALKBH5 to eliminate site-specific m6A modifications. Setting the m6A modification on the 3´ UTR of actin beta (ACTB) mRNA leads to RNA degradation, and removing the m6A modification at A2577 of Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) leads to structural changes and alters the interaction with the RNA-binding protein hnRNPC [Citation87,Citation88]. m6A modification of the RRACH motif of PVT1 by ALKBH5 promotes the proliferation and invasive metastasis of osteosarcoma cells [Citation59]. The m6A motif of WTAP is GGACU, which matches the candidate m6A modification site of FOXD2-AS1 and improves FOXD2-AS1 stability, which in turn accelerates the progression of osteosarcoma [Citation54]. Therefore, applying CRISPR technology to target common sequences where m6A bindings to ncRNAs increases or decreases the level of m6A modified scripts, affecting downstream target genes and regulations of the biological functions of osteosarcoma cells. As potential targets, ncRNAs offer new possibilities for the clinical treatment of osteosarcoma through association with m6A modifications.
Conclusion and perspectives
Over the past decades, m6A modifications have been demonstrated to be present in DNA, RNA, and proteins and have been associated with the biological properties of a variety of malignant tumours, such as gastric cancer, colon cancer, lung cancer, and osteosarcoma [Citation89,Citation90]. In osteosarcoma, dysregulation of global m6A levels and the expression of m6A regulators (writers, erasers, and readers) may be associated with metastasis, drug resistance, and prognosis in osteosarcoma patients [Citation15]. Benefiting from emerging technologies and bioinformatics analysis, an increasing number of m6A-modified ncRNAs have been identified, annotated, and functionally predicted. In osteosarcoma, modification of miRNAs by m6A mainly regulates their maturation process, modification of lncRNAs regulates stability and degradation, and modification of circRNAs can affect the circRNA-miRNA-mRNA axis. ncRNAs can also modulate m6A modification during posttranscriptional regulation and mediate the expression of m6A regulators. ncRNAs can bind to m6A regulators and participate in the interaction of m6A regulators with their target RNAs.
Notably, METTL14 and ALKBH5 have been reported as oncogenes and suppressor genes in osteosarcoma, which is transitional. This phenomenon has also been reported in other cancers. In colorectal cancer, METTL3 enhances MYC expression in an m6A IGF2BP1-dependent manner, while in another study, METTL3 modified the p38/ERK pathways and played a tumour suppressive role [Citation91,Citation92]. The dual roles of m6A regulators may be due to the different origins of tumour tissues, and tumour tissues with different grades and stages may have different expression levels. Meanwhile, m6A binds to different downstream targets, and the functional impact on downstream processes may be highly heterogeneous [Citation93]. In addition, m6A modification is a dynamic and reversible process, and different detection time points may lead to inconsistent results. m6A regulators act in tumours through different mechanisms of action, and more convincing mechanisms of action need to be explored.
Although m6A modification is widely present in various RNA types, most studies have focused only on its presence in mRNAs, and there are still many m6A-modified ncRNAs that have yet to be identified. In addition to common miRNAs, circRNAs, lncRNAs, ncRNAs also include Small nuclear RNAs (snRNAs), Small nucleolar RNAs (snoRNAs), and PIWI-interacting RNAs (piRNAs) [Citation94], and the effects of m6A interactions with these ncRNAs on the development of osteosarcoma have not yet been reported, and further studies are needed to confirm them. With the rapid development of bioinformatics analysis and gene sequencing technology, more and more m6A modification sites of ncRNAs will be identified. Specific phenotypic alterations may not be exclusively caused by alterations in a single type of RNA modification. RNA modifications such as N1-methyladenosine (m1A), 5-methylcytosine (m5C), 7-methylguanosine (m7G), and 2’-O-methylation (Nm) may also act concurrently with m6A modifications on ncRNA [Citation95]. The diagnostic sensitivity and specificity of m6A-associated ncRNAs as potential tumour markers need further validation, clinical diagnostic and therapeutic approaches are lacking, and more in-depth explorations are necessary to determine their reference ranges in body fluids.
Overall, this paper provides a systematic review of the biological functions of m6A regulators in osteosarcoma and investigates the interaction between m6A modifications and ncRNAs in osteosarcoma. The study of m6A and ncRNA interregulation provides a new direction to study the pathogenesis of osteosarcoma. As research progresses, more molecular regulatory mechanisms between ncRNA and m6A will be discovered in osteosarcoma, and targeting m6A-modified ncRNAs will be a promising therapeutic approach.
Authors’ contributions
YW contributed to the concept and design of the paper. YZ planned and wrote the paper. YZ, YX, YL, GQ, MH, and JL contributed to generating the Figures and Tables. YB participated in the revision of the Figures and Tables. BC participated in the summary of the paper. YW and JX participated in the revision of the paper. All authors read and approved the final paper.
Consent for publication
All participants were informed and gave written consent.
Availability of data and material
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Simpson E, Brown HL. Understanding osteosarcomas. JAAPA. 2018;31(8):15–17. doi: 10.1097/01.JAA.0000541477.24116.8d
- Broadhead ML, Clark JCM, Myers DE, et al. The molecular pathogenesis of osteosarcoma: a review. Sarcoma. 2011;2011:1–12. doi: 10.1155/2011/959248
- Ping XL, Sun B-F, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24(2):177–189. doi: 10.1038/cr.2014.3
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112
- Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation Reveals Enrichment in 3´ UTRs and near stop codons. Cell. 2012;149149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003
- Coker H, Wei G, Brockdorff N. m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim Biophys Acta, Gene Regul Mech. 2019;1862(3):310–318. doi: 10.1016/j.bbagrm.2018.12.002
- Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi: 10.1038/nrc.2017.99
- Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16(3):167–179. doi: 10.1038/nrd.2016.117
- Chen Y, Lin Y, Shu Y, et al. Interaction between N(6)-methyladenosine (m(6)A) modification and noncoding RNAs in cancer. Mol Cancer. 2020;19(1):94. doi: 10.1186/s12943-020-01207-4
- Liu N, Dai Q, Zheng G, et al. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature. 2015;518(7540):560–564. doi: 10.1038/nature14234
- Shen S, Zhang R, Jiang Y, et al. Comprehensive analyses of m6A regulators and interactive coding and non-coding RNAs across 32 cancer types. Mol Cancer. 2021;20(1):67. doi: 10.1186/s12943-021-01362-2
- Li Y, Xiao J, Bai J, et al. Molecular characterization and clinical relevance of m(6)A regulators across 33 cancer types. Mol Cancer. 2019;18(1):137. doi: 10.1186/s12943-019-1066-3
- Ma JZ, Yang F, Zhou C-C, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65(2):529–543. doi: 10.1002/hep.28885
- Ma S, Chen C, Ji X, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12(1):121. doi: 10.1186/s13045-019-0805-7
- Yang X, Hu X, Liu J, et al. N6-methyladenine modification in noncoding RNAs and its function in cancer. Biomark Res. 2020;8(1):61. doi: 10.1186/s40364-020-00244-x
- Li HB, Huang G, Tu J, et al. METTL14-mediated epitranscriptome modification of MN1 mRNA promote tumorigenicity and all-trans-retinoic acid resistance in osteosarcoma. EBioMedicine. 2022;82:104142. doi: 10.1016/j.ebiom.2022.104142
- Yuan Y, Yan G, He M, et al. ALKBH5 suppresses tumor progression via an m(6)A-dependent epigenetic silencing of pre-miR-181b-1/YAP signaling axis in osteosarcoma. Cell Death Dis. 2021;12(1):60. doi: 10.1038/s41419-020-03315-x
- Liu J, Yue Y, Han D, et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–95. doi: 10.1038/nchembio.1432
- Deng X, Su R, Weng H, et al. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28(5):507–517. doi: 10.1038/s41422-018-0034-6
- Schwartz S, Mumbach M, Jovanovic M, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5´ sites. Cell Rep. 2014;8(1):284–296. doi: 10.1016/j.celrep.2014.05.048
- Warda AS, Kretschmer J, Hackert P, et al. Human METTL16 is a N 6 -methyladenosine (m 6 A) methyltransferase that targets pre-mRnas and various non-coding RNAs. EMBO Rep. 2017;18(11):2004–2014. doi: 10.15252/embr.201744940
- Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687
- Zheng G, Dahl J, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015
- Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74(4):640–650. doi: 10.1016/j.molcel.2019.04.025
- Haussmann IU, Bodi Z, Sanchez-Moran E, et al. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust drosophila sex determination. Nature. 2016;540(7632):301–304. doi: 10.1038/nature20577
- Meyer KD, Jaffrey SR. Rethinking m(6)A readers, writers, and erasers. Annu Rev Cell Dev Biol. 2017;33:319–342. doi: 10.1146/annurev-cellbio-100616-060758
- Huang H, Weng H, Sun W, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285–295. doi: 10.1038/s41556-018-0045-z
- Li J, Rao B, Yang J, et al. Dysregulated m6A-Related regulators are associated with tumor metastasis and poor prognosis in osteosarcoma. Front Oncol. 2020;10:769. doi: 10.3389/fonc.2020.00769
- Zhang W, Wang L, Zhang P, et al. m6A regulators are associated with osteosarcoma metastasis and have prognostic significance: a study based on public databases. Medicine (Baltimore). 2021;100(20):e25952. doi: 10.1097/MD.0000000000025952
- Wang Y, Zeng L, Liang C, et al. Integrated analysis of transcriptome-wide m 6 a methylome of osteosarcoma stem cells enriched by chemotherapy. Epigenomics. 2019;11(15):1693–1715. doi: 10.2217/epi-2019-0262
- Huang H, Weng H, Chen J. m(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37(3):270–288. doi: 10.1016/j.ccell.2020.02.004
- He L, Li H, Wu A, et al. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18(1):176. doi: 10.1186/s12943-019-1109-9
- Zhang W, He L, Liu Z, et al. Multifaceted functions and novel insight into the regulatory role of RNA N(6)-methyladenosine modification in musculoskeletal disorders. Front Cell Dev Biol. 2020;8:870. doi: 10.3389/fcell.2020.00870
- Ling Z, Chen L, Zhao J. m6A-dependent up-regulation of DRG1 by METTL3 and ELAVL1 promotes growth, migration, and colony formation in osteosarcoma. Biosci Rep. 2020;40(4). doi: 10.1042/BSR20200282
- Wang J, Wang W, Huang X, et al. m6A-dependent upregulation of TRAF6 by METTL3 is associated with metastatic osteosarcoma. J Bone Oncol. 2022;32:100411. doi: 10.1016/j.jbo.2022.100411
- Miao W, Chen J, Jia L, et al. The m6A methyltransferase METTL3 promotes osteosarcoma progression by regulating the m6A level of LEF1. Biochem Biophys Res Commun. 2019;516(3):719–725. doi: 10.1016/j.bbrc.2019.06.128
- Wei K, Gao Y, Wang B, et al. Methylation recognition protein YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) regulates the proliferation, migration and invasion of osteosarcoma by regulating m6A level of CCR4-NOT transcription complex subunit 7 (CNOT7). Bioengineered. 2022;13(3):5236–5250.
- Zhou C, Zhang Z, Zhu X, et al. N6-methyladenosine modification of the TRIM7 positively regulates tumorigenesis and chemoresistance in osteosarcoma through ubiquitination of BRMS1. EBioMedicine. 2020;59:102955.
- Wang C, Meng Y, Zhao J, et al. Deubiquitinase USP13 regulates glycolytic reprogramming and progression in osteosarcoma by stabilizing METTL3/m 6 A/ATG5 axis. Int J Biol Sci. 2023;19(7):2289–2303. doi: 10.7150/ijbs.82081
- Chen S, Li Y, Zhi S, et al. WTAP promotes osteosarcoma tumorigenesis by repressing HMBOX1 expression in an m(6)A-dependent manner. Cell Death Dis. 2020;11(8):659. doi: 10.1038/s41419-020-02847-6
- Liu Z, Liu N, Huang Z, et al. METTL14 Overexpression promotes osteosarcoma Cell apoptosis and slows tumor progression via caspase 3 activation. Cancer Manag Res. 2020;12:12759–12767. doi: 10.2147/CMAR.S284273
- Cheng J, Xu Z, Tan W, et al. METTL16 promotes osteosarcoma progression by downregulating VPS33B in an m 6 A-dependent manner. J Cell Physiol. 2023. doi:10.1002/jcp.31068.
- Luo J, Wang X, Chen Z, et al. The role and mechanism of JAK2/STAT3 signaling pathway regulated by m6A methyltransferase KIAA1429 in osteosarcoma. J Bone Oncol. 2023;39:100471. doi: 10.1016/j.jbo.2023.100471
- Lv D, Ding S, Zhong L, et al. M(6)A demethylase FTO-mediated downregulation of DACT1 mRNA stability promotes Wnt signaling to facilitate osteosarcoma progression. Oncogene. 2022;41(12):1727–1741. doi: 10.1038/s41388-022-02214-z
- Shan HJ, Gu W X, Duan G, et al. Fat mass and obesity associated (FTO)-mediated N6-methyladenosine modification of Krüppel-like factor 3 (KLF3) promotes osteosarcoma progression. Bioengineered. 2022;13(4):8038–8050.
- Yadav P, Subbarayalu P, Medina D, et al. M6A RNA methylation regulates histone ubiquitination to Support cancer growth and progression. Cancer Res. 2022;82(10):1872–1889. doi: 10.1158/0008-5472.CAN-21-2106
- Yang Z, Cai Z, Yang C, et al. ALKBH5 regulates STAT3 activity to affect the proliferation and tumorigenicity of osteosarcoma via an m6A-YTHDF2-dependent manner. EBioMedicine. 2022;80:104019. doi: 10.1016/j.ebiom.2022.104019
- Liu D, Li Z, Zhang K, et al. N(6)-methyladenosine reader YTHDF3 contributes to the aerobic glycolysis of osteosarcoma through stabilizing PGK1 stability. J Cancer Res Clin Oncol. 2023;149(8):4601–4610. doi: 10.1007/s00432-022-04337-y
- He Q, Hao P, He G, et al. IGF2BP1-regulated expression of ERRα is involved in metabolic reprogramming of chemotherapy resistant osteosarcoma cells. J Transl Med. 2022;20(1):348. doi: 10.1186/s12967-022-03549-7
- Pan Y, Ma P, Liu Y, et al. Multiple functions of m(6)A RNA methylation in cancer. J Hematol Oncol. 2018;11(1):48. doi: 10.1186/s13045-018-0590-8
- Zhou X, Yang Y, Li Y, et al. METTL3 contributes to osteosarcoma progression by increasing DANCR mRNA stability via m6A modification. Front Cell Dev Biol. 2021;9:784719. doi: 10.3389/fcell.2021.784719
- Wang X, Feng J, Xue Y, et al. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature. 2016;534(7608):575–578. doi: 10.1038/nature18298
- Liu H, Shu W, Liu T, et al. Analysis of the function and mechanism of DIRAS1 in osteosarcoma. Tissue Cell. 2022;76:101794. doi: 10.1016/j.tice.2022.101794
- Ren Z, Hu Y, Sun J, et al. N(6)-methyladenosine methyltransferase WTAP-stabilized FOXD2-AS1 promotes the osteosarcoma progression through m(6)A/FOXM1 axis. Bioengineered. 2022;13(4):7963–7973.
- Xie Y, Castro-Hernández R, Sokpor G, et al. RBM15 Modulates the function of Chromatin Remodeling factor BAF155 through RNA methylation in developing cortex. Mol Neurobiol. 2019;56(11):7305–7320. doi: 10.1007/s12035-019-1595-1
- Yang F, Liu Y, Xiao J, et al. Circ-CTNNB1 drives aerobic glycolysis and osteosarcoma progression via m6A modification through interacting with RBM15. Cell Prolif. 2022;56(1):e13344. doi: 10.1111/cpr.13344
- Doxtader KA, Wang P, Scarborough AM, et al. Structural basis for regulation of METTL16, an S-Adenosylmethionine homeostasis factor. Mol Cell. 2018;71(6):1001–1011.e4. doi: 10.1016/j.molcel.2018.07.025
- Zhu W, Wang J-Z, Wei J-F, et al. Role of m6A methyltransferase component VIRMA in multiple human cancers (review). cancer Cell int. Cancer Cell Int. 2021;21(1):172. doi: 10.1186/s12935-021-01868-1
- Chen S, Zhou L, Wang Y. ALKBH5-mediated m(6)A demethylation of lncRNA PVT1 plays an oncogenic role in osteosarcoma. Cancer Cell Int. 2020;20:34. doi: 10.1186/s12935-020-1105-6
- Shi R, Ying S, Li Y, et al. Linking the YTH domain to cancer: the importance of YTH family proteins in epigenetics. Cell Death Dis. 2021;12(4):346. doi: 10.1038/s41419-021-03625-8
- Cao D, Ge S, Li M. MiR-451a promotes cell growth, migration and EMT in osteosarcoma by regulating YTHDC1-mediated m6A methylation to activate the AKT/mTOR signaling pathway. J Bone Oncol. 2022;33:100412. doi: 10.1016/j.jbo.2022.100412
- Terasaka T, Kim T, Dave H, et al. The RNA-Binding protein ELAVL1 regulates GnRH receptor expression and the response to GnRH. Endocrinology. 2019;160(8):1999–2014. doi: 10.1210/en.2019-00203
- Zheng H, Wang J, Zhang W, et al. Mechanism for Bioactive Nanomaterial circ0024831 regulation of staphylococcal nuclease domain containing 1 via RNA methylation recognition in osteosarcoma. J Biomed Nanotechnol. 2022;18(2):453–462. doi: 10.1166/jbn.2022.3256
- Yang J, Han Q, Li C, et al. circular RNA circ_0001105 inhibits progression and metastasis of osteosarcoma by sponging miR-766 and activating YTHDF2 expression. Onco Targets Ther. 2020;13:1723–1736. doi: 10.2147/OTT.S234668
- Meng Y, Hao D, Huang Y, et al. Circular RNA circNRIP1 plays oncogenic roles in the progression of osteosarcoma. Mamm Genome. 2021;32(6):448–456. doi: 10.1007/s00335-021-09891-3
- Beermann J, Piccoli M-T, Viereck J, et al. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. doi: 10.1152/physrev.00041.2015
- Han J, Lee Y, Yeom K H, et al. Molecular basis for the recognition of primary microRnas by the drosha-DGCR8 complex. Cell. 2006;125(5):887–901. doi: 10.1016/j.cell.2006.03.043
- Chen T, Hao Y-J, Zhang Y, et al. m(6)A RNA methylation is regulated by microRnas and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16(3):289–301. doi: 10.1016/j.stem.2015.01.016
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi: 10.1038/nbt.2890
- Chen RX, Chen X, Xia L-P, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10(1):4695. doi: 10.1038/s41467-019-12651-2
- Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31
- Lee Y, Choe J, Park O H, et al. Molecular mechanisms driving mRNA degradation by m(6)A modification. Trends Genet. 2020;36(3):177–188.
- Huang R, Zhang Y, Bai Y, et al. N6-methyladenosine modification of fatty acid amide hydrolase Messenger RNA in circular RNA STAG1–regulated astrocyte dysfunction and depressive-like behaviors. Biol Psychiatry. 2020;88(5):392–404. doi: 10.1016/j.biopsych.2020.02.018
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993
- Chi Y, Wang D, Wang J, et al. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;8(9):1015.
- Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028
- Patil DP, Chen C-K, Pickering BF, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537(7620):369–373. doi: 10.1038/nature19342
- Derderian C, Orunmuyi AT, Olapade-Olaopa EO, et al. PVT1 signaling is a Mediator of cancer progression. Front Oncol. 2019;9:502. doi: 10.3389/fonc.2019.00502
- Gu Y, Wu X, Zhang J, et al. The evolving landscape of N(6)-methyladenosine modification in the tumor microenvironment. Mol Ther. 2021;29(5):1703–1715.
- Bi Y, Meng D, Wan M, et al. m6A-Related lncRnas predict Overall survival of patients and regulate the tumor immune microenvironment in osteosarcoma. Comput Intell Neurosci. 2022;2022:1–16. doi: 10.1155/2022/9315283
- Wang S, Ma F, Feng Y, et al. Role of exosomal miR‑21 in the tumor microenvironment and osteosarcoma tumorigenesis and progression (review). Int J Oncol. 2020;56(5):1055–1063. doi: 10.3892/ijo.2020.4992
- Jiang J, Qu H, Zhan X, et al. Identification of osteosarcoma m6A-related prognostic biomarkers using artificial intelligence: RBM15. Sci Rep. 2023;13(1):5255. doi: 10.1038/s41598-023-28739-1
- Cacabelos R. Parkinson’s disease: from pathogenesis to pharmacogenomics. Int J Mol Sci. 2017;18(3):551. doi: 10.3390/ijms18030551
- Zhou K, Sun Y, Dong D, et al. EMP3 negatively modulates breast cancer cell DNA replication, DNA damage repair, and stem-like properties. Cell Death Dis. 2021;12(9):844. doi: 10.1038/s41419-021-04140-6
- Salehan MR, Morse HR. DNA damage repair and tolerance: a role in chemotherapeutic drug resistance. Br J Biomed Sci. 2013;70(1):31–40. doi: 10.1080/09674845.2013.11669927
- Xiang Y, Laurent B, Hsu C-H, et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543(7646):573–576. doi: 10.1038/nature21671
- Liu XM, Zhou J, Mao Y, et al. Programmable RNA N(6)-methyladenosine editing by CRISPR-Cas9 conjugates. Nat Chem Biol. 2019;15(9):865–871. doi: 10.1038/s41589-019-0327-1
- Chen M, Wong CM. The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Mol Cancer. 2020;19(1):44.
- Zeng C, Huang W, Li Y, et al. Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J Hematol Oncol. 2020;13(1):117. doi: 10.1186/s13045-020-00951-w
- Ianniello Z, Paiardini A, Fatica A. N(6)-methyladenosine (m(6)A): a promising new molecular target in acute myeloid leukemia. Front Oncol. 2019;9:251. doi: 10.3389/fonc.2019.00251
- Xiang S, Liang X, Yin S, et al. N6-methyladenosine methyltransferase METTL3 promotes colorectal cancer cell proliferation through enhancing MYC expression. Am J Transl Res. 2020;12(5):1789–1806.
- Deng R, Cheng Y, Ye S, et al. m6A methyltransferase METTL3 suppresses colorectal cancer proliferation and migration through p38/ERK pathways. Onco Targets Ther. 2019;12:4391–4402. doi: 10.2147/OTT.S201052
- Zhang Z, Luo K, Zou Z, et al. Genetic analyses support the contribution of mRNA N(6)-methyladenosine (m(6)A) modification to human disease heritability. Nat Genet. 2020;52(9):939–949. doi: 10.1038/s41588-020-0644-z
- Wajahat M, Bracken CP, Orang A. Emerging functions for snoRnas and snoRNA-Derived fragments. Int J Mol Sci. 2021;22(19):10193. doi: 10.3390/ijms221910193
- Zhang T, Gu J, Wang X, et al. RNA methylation regulators contribute to poor prognosis of hepatocellular carcinoma associated with the suppression of bile acid metabolism: a multi-omics analysis. Am J Cancer Res. 2022;12(7):2989–3013.
