ABSTRACT
Currently, clinicians use their judgement and indices such as the Prediction of Alcohol Withdrawal Syndrome Scale (PAWSS) to determine whether patients are admitted to hospitals for consideration of withdrawal syndrome (AWS). However, only a fraction of those admitted will experience severe AWS. Previously, we and others have shown that epigenetic indices, such as the Alcohol T-Score (ATS), can quantify recent alcohol consumption. However, whether these or other alcohol biomarkers, such as carbohydrate deficient transferrin (CDT), could identify those at risk for severe AWS is unknown. To determine this, we first conducted genome-wide DNA methylation analyses of subjects entering and exiting alcohol treatment to identify loci whose methylation quickly reverted as a function of abstinence. We then tested whether methylation at a rapidly reverting locus, cg07375256, or other existing metrics including PAWSS scores, CDT levels, or ATS, could predict outcome in 125 subjects admitted for consideration of AWS. We found that PAWSS did not significantly predict severe AWS nor seizures. However, methylation at cg07375256 (ZSCAN25) and CDT strongly predicted severe AWS with ATS (p < 0.007) and cg07375256 (p < 6 × 10–5) methylation also predicting AWS associated seizures. We conclude that epigenetic methods can predict those likely to experience severe AWS and that the use of these or similar Precision Epigenetic approaches could better guide AWS management.
Introduction
Between 2014 and 2018, alcohol use disorder (AUD) was present in 1 in 11 emergency room (ER) visits in the United States [Citation1,Citation2]. In each of these alcohol-related ER presentations, clinicians must assess potential for Alcohol Withdrawal Syndrome (AWS) with nearly 1.4 million of these ER patients being admitted in the hopes of preventing severe AWS.
AWS is a syndrome of autonomic dysregulation, neurologic and psychiatric signs, and symptoms that can begin a few hours or several days after a reduction or cessation in drinking [Citation3]. Although generally mild, severe AWS can be fatal with hallucinations, delirium, and seizures being frequently observed [Citation4]. Unfortunately, there is no generally accepted methods for determining who is at risk for AWS with only a minority of the admitted patients, between 2% and 7%, actually developing severe AWS [Citation5,Citation6]. As a result, clinicians tend to err on the side of caution and admit many patients who could otherwise be managed in less restrictive care settings.
According to the American Society of Addiction Medicine (ASAM), the use of clinician administered scales such as The Prediction of Alcohol Withdrawal Severity Scale (PAWSS) or the Lübeck Alcohol withdrawal Risk Scale (LARS) can be used for assessing risk of severe AWS [Citation7,Citation8]. However, in their review of 530 studies of AWS, Wood and colleagues noted a potential for bias and a lack of independent validation of these AWS prediction scales [Citation6]. Hence, there is a strong need for either independent validation of the clinical utility of these tools, or if not useful, the derivation of new measures capable of helping clinicians decide whether to admit a prospective patient for more intensive monitoring and/or treatment.
Advances in our understanding of the effect of alcohol on the methylome may provide a new measure to predict AWS. Specifically, in prior work, we have shown that a metric composed of the unweighted Z-scores of the results from four methylation sensitive digital PCR assays (MSdPCR), referred to as the Alcohol T Score (ATS), is strongly correlated with other objective biomarkers of heavy alcohol consumption (HAC) and accurately predicted those who were admitted for residential treatment of AUD [Citation9,Citation10]. However, whereas this tool may be useful for predicting both chronic HAC and the biological consequences of HAC [Citation11,Citation12], the dynamic response time of the four loci is slow, with a half-time of reversion of the most responsive locus, cg04987734, being on the order of 3 months [Citation9]. Whereas this slow dynamic may make the ATS useful for detecting chronic HAC, it may make the ATS less useful for other purposes. For example, if the vulnerability to AWS can develop in less time, the ATS may lack the required sensitivity for use as a useful clinical tool for predicting AWS.
Clinicians have long sought a useful tool for predicting AWS. But at the current time, there are at least two key barriers for training and testing a better method for predicting severe AWS. The first barrier is that the minimum amount and timing of alcohol consumption necessary to establish vulnerability to AWS is not known. In the early 1950s, Isbell and colleagues enrolled 10 subjects in a study in which the subjects each ingested between 286 and 489 ml of 95% ethanol, based on body weight, for at least 7 and up to 87 days [Citation13]. The four subjects who drank 34 days or less experienced clear signs of what we now term AWS but did not experience seizures or delirium tremens. However, of the six subjects who drank between 48 and 87 days, two had seizures and three had frank delirium. Similarly, in the early 1960s, Mendelson and Ladou enrolled 10 subjects with ‘long histories of alcoholism’ but no history of seizures who had been abstinent for between 10 and 37 days, into a protocol where the subjects ingested an average of 30 ounces of 86 proof alcohol for 24 consecutive days, then were monitored signs of alcohol withdrawal [Citation14]. Although numerous symptoms of AWS were noted to be present in these subjects, no seizures nor frank delirium were observed. Critically, both sets of investigators took pains to ensure their subjects had adequate nutrition, and it is important to note that the use of other substances that might affect the likelihood of severe AWS were not permitted [Citation15]. Therefore, although these older studies have significant limitations, they do indicate that a sustained period of heavy drinking for at least three to 4 weeks is necessary to induce vulnerability to AWS.
The second barrier to developing better tool for predicting AWS is that even if we could establish the minimum dose and timing of alcohol necessary to become vulnerable to AWS, the ability of patients to accurately report their prior alcohol intake in clinical settings is less than optimal [Citation16,Citation17]. This problem is particularly exacerbated in the ER setting where involuntary hospitalization for AWS is being considered. Therefore, any clinical tool solely based on self-report may have a high failure rate.
Potentially, a currently existing biomarker of alcohol use that captures the amount and chronicity of alcohol consumption relevant to AWS could be used to predict the likelihood of AWS. Specifically, both phosphatidyl ethanolamine (PEth) and carbohydrate deficient transferrin (CDT) assessments are now used in some clinical settings to assess recent alcohol use [Citation18,Citation19]. However, in 34 patients admitted for treatment of AWS, Novak and colleagues found that PEth levels were only modestly associated with severity of AWS and did not predict severe AWS [Citation20]. Furthermore, Helander and associates note considerable variation in PEth catabolism that may make it less suitable as biomarker for predicting AWS [Citation21,Citation22]. Similarly, examinations of the CDT have shown only modest power of the assay to predict severe AWS [Citation23,Citation24]. Therefore, although the ability of these biomarkers to predict more recent alcohol intake is not in question, it seems unlikely that either of these two biomarkers could serve as useful clinical tools for predicting AWS.
One reason for the failure of these two biomarkers to predict AWS may be the relatively short half-life of the two biomarkers. PEth has a elimination half-life of 4–10 days [Citation21]. The CDT has half-life between 7 and 10 days [Citation21,Citation25]. If the window for assessing alcohol consumption needs to be longer, as suggested by the earlier studies of Isbell and colleagues, these assays may not be able to capture use during the period of time critical to the induction of seizure vulnerability.
In 2019, we began a National Institutes of Health (NIH) Small Business Innovation Research (SBIR) project to identify methylation markers with reversion half-lives considerably less than those contained in the ATS. In this communication, we report on the development of those markers using DNA and biomaterial from a previously conducted study. We then describe a prospective study of 125 subjects that tested the ability of an MSdPCR assay developed from that effort, along with the PAWSS, the ATS, and CDT, to predict features of severe AWS.
Materials and methods
Human subject methods for methylation locus discovery
The identification of rapidly reverting methylation loci used clinical data and biomaterials collected from a study conducted between 2016 and 2018. This effort, which collected subjects with HAC as they entered and exited alcohol treatment, has been previously described with all protocols and procedures being approved by the Western Institutional Review Board (WIRB 20,160,135) [Citation9,Citation26]. In brief, these subjects were recruited from one of the three Iowa inpatient treatment facilities near Iowa City in the years 2016–2018. Each of these facilities offered detoxification services followed by a 21 to 28-day inpatient alcohol treatment program. Any individual who: 1) was over the age of 18, 2) was capable of giving consent in English, 3) who was admitted to one of these facilities for treatment of current sustained alcohol dependence in the context of current alcohol intoxication and 4) expressed interest in our protocol to a member of their healthcare team was eligible to participate in the study. Potential subjects were not approached for intake into the study if they were still intoxicated or judged to be under the influence of other substances. After written informed consent for the study was obtained, each participant was interviewed with a series of instruments including the Substance Use Questionnaire [Citation27]. After the interview process was complete, each participant was then given a contact card and urged to contact the facility staff or the research assistant if they were interested in completing the exit interview that was held as close to the discharge date as possible. Fifty four of the 143 individuals who participated in the intake interview also completed the discharge interview, which consisted of an updated Substance Use Questionnaire and phlebotomy.
Control subjects for this study were recruited from the University of Iowa via email. Any subject over the age of 21 who denied any use of alcohol or illegal substances, including cannabis, in the past year and denied a history of past substance abuse, except for tobacco use disorder, was eligible. After consent was obtained, these subjects were interviewed with the same instruments as the case subjects and then phlebotomized to provide biomaterials for the study. A total of 210 participants enrolled in the control arm of the study successfully provided clinical and biomaterial for the study with a subset of 47 of these samples being used for this study. DNA for these and the above HAC subjects were prepared as previously described [Citation9].
Methylation data for the identification of quickly reverting loci was conducted using intake (Time 1 or T1) and discharge (T2) samples from 47 individuals with HAC described above. After preparation, genome-wide methylation assessments using the Infinium MethylationEpic Version 1.0 were conducted by the Mayo Clinic Genomic lab (Rochester MN). The resulting data were then processed and cleaned using our normal procedures with a total of 824,807 probes for 45 pairs of DNA samples’ surviving quality control [Citation28–32]. The resulting M-values for these samples were then converted to beta values. The significance of methylation change between T1 and T2 at each of the loci was then evaluated using Student’s T-Test, with the subsequent p-values adjusted for multiple comparisons by Bonferonni correction [Citation33,Citation34].
The sequence surrounding the primary candidate locus, cg07375256, was then downloaded from the UCSC Genome browser. Fluorescent methylation sensitive digital PCR primer probe sets were then generated for the locus using our proprietary methods as previously described [Citation9].
Human subject methods for collecting AWS test cohort
The Alcohol Withdrawal Syndrome (AWS) subjects were patients who were admitted to the University of Iowa Hospital between May 2022 and May 2023 for the management of alcohol withdrawal. All protocols and procedures used were approved by the University of Iowa Institutional Review Board (IRB 202,103,577). In brief, after detoxifying, subjects who expressed interest were approached by staff members and educated on the protocol, and if still interested, enrolled in the study. After written informed consent was received, subjects were interviewed with a REDCap administered battery including the PAWSS, a modified version of the Substance Use Questionnaire and a battery of AUD-related modules from the PhenX project [Citation35], then phlebotomized for preparation of DNA samples and serum CDT assays. Key outcome variables were ascertained through chart review by a University of Iowa psychiatry resident with experience in the treatment of AUD (BP). Each clinical chart review was then inspected by a University of Iowa faculty psychiatrist (AA). Both reviewers were blind to biological measure outcomes, and the study team had no role in choosing the treatment of study subjects. Clinical outcomes ascertained included the occurrence of seizures, hallucinations, amount of benzodiazepines (BZD) administered, administration of phenobarbital-in the case of severe AWS, and whether the use of the word ‘delirium tremens’ was used as a diagnosis in the medical record. Conversion of BZD dose to diazepam equivalents was done according to the method of Salzman and colleagues [Citation36].
Control subjects were recruited from the University of Iowa community using emails targeting abstinent or non-abstinent individuals. After informed consent was received, these subjects were also interviewed with a REDCap administered battery including the Michigan Alcohol Screening Test (MAST), a modified version of the Substance Use Questionnaire, and a health history questionnaire, then phlebotomized [Citation7,Citation27,Citation35].
ATS levels and MSdPCR assessment of cg07375256 levels were conducted using reagents and software from Behavioral Diagnostics (Coralville, IA) and from Bio-Rad (Hercules, CA) using our previously published methods [Citation9,Citation10,Citation12,Citation37]. Similarly, the smoking intensity at cg05575921, a generally accepted biomarker for smoking intensity [Citation38], was assessed using reagents and software from Behavioral Diagnostics (Coralville, IA) and from Bio-Rad (Hercules, CA) according to our previously described methods [Citation11].
Carbohydrate Deficient Transferrin levels were determined by Clinical Reference Laboratories (https://www.crlcorp.com/, Lenexa, KS) using capillary electrophoresis [Citation39].
Data were analysed in R [Citation40] using standard chi-squared tests, analysis of variance, and Kruskal-Wallis tests, as appropriate. To assess the utility of the ATS, Dcg07375256, and the PAWSS in predicting clinical outcomes, receiver operating characteristic (ROC) area under the curve (AUC) statistics were computed with the confidence interval (CI) estimated by non-parametric bootstrap using the pROC package [Citation41]. Except where indicated, all p-values reported are nominal.
Results
gives the clinical characteristics of the 45 Locus Discovery subjects collected between the years 2016–2018 whose entry (T1) and exit (T2) DNA methylation values were used to identify quickly reverting DNA methylation loci. In brief, these HAC subjects tended to be in their early 40s with approximately 90% of the sample being European American (40 of 45). As per our 2019 publication, these subjects had all reported drinking almost every day for the 8 weeks prior admission and ingested approximately 15 standard drinks per day in the month prior to admission. Their first blood sample (T1) was taken between 2 and 7 days after admission to the facility. The second sample was taken shortly before the study's exit approximately 3 weeks after the T1 sampling.
Table 1. Clinical characteristics of the subjects whose data were used to identify candidate markers.
The genome-wide methylation values from the 45 paired DNA samples were evaluated with respect to two criteria. The first criterion was that the changes in DNA methylation be statistically significant. After Bonferroni correction, methylation at 18 loci were significantly different between intake and exit time points (see supplemental Table S1). The second criterion was that the absolute difference in beta values between the T1 and T2 timepoints be at least 5%. This is because MSdPCR values have a fixed lower limit of precision and we wished to maximize signal-to-noise ratios. Of the 824,807 probes tested, seven had changes ≥ 4.7% () lists the seven loci that had an absolute difference of DNA methylation values of 4.7%. Although all seven of these loci had changes in methylation that were nominally different, after Bonferroni correction, only one locus, cg09807356 had significantly different between study entry and exit.
Table 2. Leading candidates from initial screening efforts to identify rapidly reverting loci.
Since five of the seven loci were also profiled in our prior examination of alcohol reversion that used a previous generation of Illumina methylation array, we examined the differences in DNA methylation between the entry and exit found in those samples as listed in Supplementary Table S2 of that 2014 study [Citation27]. Interestingly, the directionality of change in methylation was the same at all five loci with cg07375256 also having an absolute difference of 5.6% between study intake and exit in that study as well suggesting that methylation at this locus reliably changed as a function of abstinence from alcohol (see ).
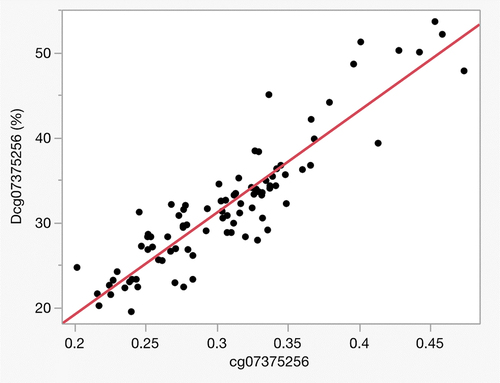
We then developed an MSdPCR for the cg07375256 locus because it had at least nominally significant changes of > 5% in both the current and 2014 studies. According to the University of Santa Cruz Genome browser, the CpG targeted by this probe maps to intron 7 of ZSCAN25 on Chromosome 7 [Citation42]. illustrates the relationship between DNA methylation at cg07375256 as determined by the Dcg07375256 MSdPCR assay (which is termed Dcg07375256 to distinguish it from the array measurements) in 45 pairs of T1 and T2 DNA samples from our 2019 study. Overall, the correlation between the methylation values derived from the array (cg07375256) with those from Dcg07375256 assay correlated well with a (r = 0.92). Consistent with the development of prior MSdPCR markers [Citation9,Citation11], the range for the Dcg07375256 values was greater than that for the Illumina array probe.
Figure 1. The correlation between the Illumina probe assessment and the MSdPCR assessments at the Alc12 locus (n = 90) using the Dcg07375256 assay. Overall, the r = 0.92 with the dynamic range of the ddPCR being considerably greater than that of the methylation array.
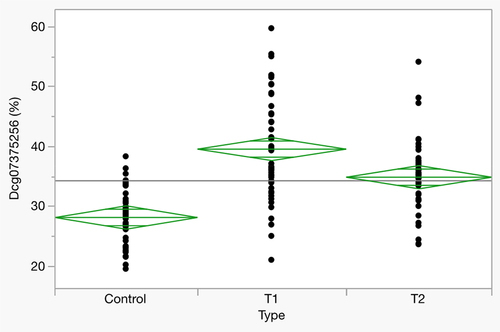
illustrates the reversion of DNA methylation in these subjects as a function of inpatient enforced abstinence from alcohol while provides the clinical characteristics of this subset of subjects from our 2019 study. At study intake (T1), subjects had an average methylation of 39.6% ± 8.8%. By 3 weeks, methylation at that site had decreased to 34.9 ± 5.8% (T1 vs T2; p < 0.0001). For reference, the average methylation at this locus in 47 abstinent controls was 28.2 ± 4.7%.
Figure 2. Methylation at cg07375256 in HAC subjects at study intake (T1) and exit (T2), and in abstinent controls (n = 46, all groups).
lists key demographic and clinical variables for the AWS subjects collected between May of 2022 and May of 2023 to test whether the PAWSS, ATS or Dcg07375256 (ZSCAN25) assay predicted severe AWS, as well as a comparison sample of community-dwelling controls collected simultaneously. Each of the subjects was phlebotomized between 1 and 6 days of admission with the average length of time from admission to blood draw being less than 2 days (1.7 ± 1.2 days). Nearly 75% of the AWS subjects were male (93 of 125) with both male and female subjects being in their mid-forties. The vast majority (94%) were European American. The self-reported rate of lifetime smoking was high with nearly 60% of subjects reporting smoking at least 100 cigarettes in their lifetime. Although a noticeable fraction (18%) of the subjects reported less than five drinks per day over the past month, nearly two-thirds (66%) reported drinking 10 drinks or more per day over the past month. In comparison, the control group was demographically similar in terms of age and ethnicity but did not show a male preponderance. Only three out of the n = 133 control subject reported consuming more than four drinks per day on average in the past month, and their MAST scores were largely below the cut-off of 5 indicating concern for excessive alcohol consumption.
Table 3. Clinical characteristics of alcohol withdrawal subjects and new controls.
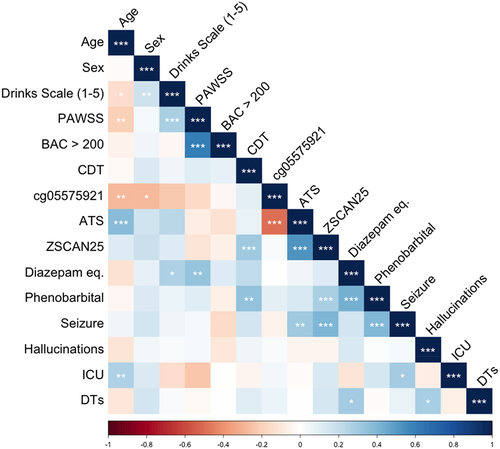
illustrates the relationships between key study variables among the AWS subjects listed in . Corresponding correlation values and significance levels are available in Supplemental Table S1. Age was significantly associated with drinks per day, PAWSS score, cg05575921 levels, and ATS values. Biological sex was associated both with self-reported drinking and cg05575921 indicated smoking intensity. Self-reported drinking was associated with age, sex, and PAWSS score. PAWSS levels were associated with age, self-reported drinking levels, having a BAC of greater than 200 mg/dl at admission and total BZD dosage. BAC levels were only associated with PAWSS scores. CDT levels were associated with Dcg07375256 (ZSCAN25) and phenobarbital administration, which is used at the University of Iowa and elsewhere to treat more severe cases of AWS [Citation43]. Dcg05575921 levels were associated with ATS scores, CDT levels, phenobarbital administration, and seizures. Diazepam dosing was associated with PAWSS and self-reported drinking levels. Phenobarbital administration was associated with CDT levels, Dcg05575921 values, and seizures. Seizures were associated with ATS scores, Dcg05575921 methylation, and phenobarbital administration. The occurrence of hallucinations was not associated with any study variable. Intensive Care Unit (ICU) admission was strongly correlated with age and weakly correlated with the occurrence of seizures. Finally, a chart diagnosis of delirium tremens (DTs) was modestly correlated with BZD dose and the presence of hallucinations.
Figure 3. The correlation of study variables to each other. * = p < 0.05, ** p < 0.01, *** p < 0.001. Age is expressed in years. Sex is binary variable whether or not the subject is male. Drinks scale is an ordinal variable with higher score indicating greater intake. PAWSS is prediction of alcohol withdrawal syndrome scale. BAC > 200 indicates if a breathalyser value greater than 200 mg/dl was noted at admission. CDT is carbohydrate transferrin levels. Cg05575921 is DNA methylation at that smoking related locus. ATS is alcohol T score. ZSCAN25 is DNA methylation per the Dcg07375256 described herein. Diazepam equivalents is the sum total equivalent of all benzodiazepines administered during hospitalization for AWS. Phenobarbital is a binomial variable indicating whether or not phenobarbital was given during hospitalization for AWS. Seizures, hallucinations, admission to the ICU, and delirium tremens (DTs) are binomial variable for the presence of those symptoms.
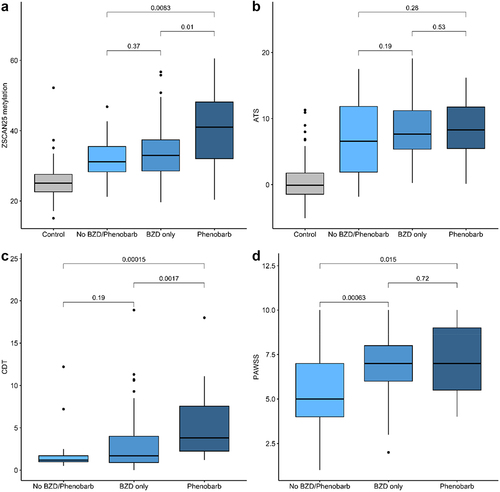
Our study examined the ability of four predictor variables, PAWSS, ATS, CDT, and Dcg07375256 (ZSCAN25), to predict key clinical outcomes. The first outcome variable was the use of medications. illustrates the relationship between each of these three predictors and one of the four treatment groups, 1) controls (not treated), 2) clinical observation of AWS subjects without the use of medications, 3) use of only BZD for the AWS subjects or 4) use of phenobarbital with or without the use of BZD for the AWS subjects. PAWSS was associated with the use of BZD but not the use of phenobarbital. ATS levels were not associated with either treatment group. CDT strongly predicted the use of Phenobarbital rather than just BZDs (Kruskal-Wallis of comparison of the use of BZDs vs the use of Phenobarbital, p < 0.002). Finally, Dcg07375256 levels were also strongly associated with the use of phenobarbital (Kruskal-Wallis of comparison of the use of BZDs vs the use of Phenobarbital, p < 0.01).
Figure 4. The relationship of predictor (PAWSS, ATS, CDT and ZSCAN (Dcg07375256)) values to treatment group. ZSCAN25 methylation and CDT values are given in percent. ATS and PAWSS have no unit values. Because control subjects were not administered the PAWSS, their values are not considered in that section of the analyses. Kruskal-Wallis p-values are given for each indicated contrasts.
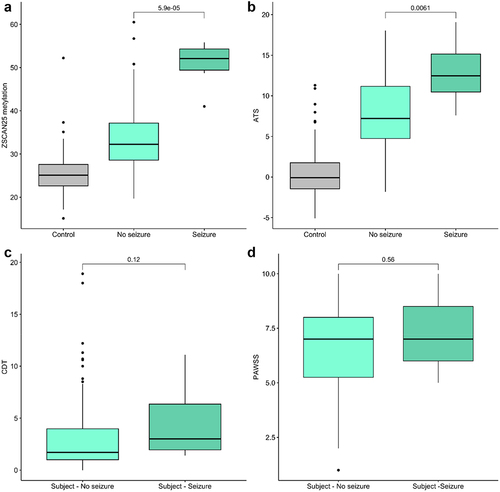
Seven alcohol-related seizures were observed among the AWS subjects during the study, including two in the BZD-only treated group and five in the phenobarbital-treated group, while none occurred in the non-medication treated group, and the difference in frequency of seizures between treatment groups was significant (chi-square = 18.37, df = 2, p < 0.001). Note that our control subjects were not clinically observed and their seizure history was not collected. illustrates the relationship of PAWSS, ATS, CDT, and Dcg07375256 values to seizures among the AWS subjects. ATS and Dcg07375256 were highly significantly associated with the occurrence of seizures (Kruskal–Wallis p < 0.001 and p < 6× 10−5). In contrast, there was no relationship of PAWSS or CDT values to the occurrence of seizures.
Figure 5. The relationship between predictor (PAWSS, ATS, CDT and ZSCAN (Dcg07375256)) levels and presence (1) or absence (0) of seizures. N = 7 for the seizure group, 116 for the no seizure group. ZSCAN25 methylation and CDT values are given in percent. ATS and PAWSS have no unit values. Kruskal-Wallis p-values are given for each indicated contrasts.
Finally, ROC AUCs were computed for the PAWSS, ATS, and Dcg07375256 as predictors of treatment group (no medication, BZD only, phenobarbital) and the occurrence of seizures. The PAWSS demonstrated fair performance (0.744, 0.607–0.866) in predicting administration of BZD vs. no treatment, but essentially no ability (0.561, 0.457–0.692) to predict the administration of phenobarbital vs. BZD only. The ATS was not able (0.569, 0.426–0.732) to predict the administration of BZD vs. no treatment, or phenobarbital vs. BZD only (0.580, 0.468, 0.721). In contrast, Dcg07375256 showed fair ability (0.737, 0.555–0.894) to predict the administration of phenobarbital vs. BZD only, but not BZD vs. no medication (0.571, 0.461–0.698). For seizures, the PAWSS showed poor ability to predict their occurrence (0.615, 0.490–0.817), whereas the ATS showed good (0.818, 0.692–0.935) and Dcg07375256 showed excellent (0.955,0.890–0.991) discrimination.
Discussion
Because it is a potentially life threatening condition, generations of clinicians have sought to develop better methods for predicting serious AWS. In this communication, we provide strong evidence that methylation at cg07375256, a CpG site in ZSCAN25, is associated with seizures and the decision to administer phenobarbital to patients who have been admitted for observation and/or treatment of AWS. As such, if replicated, these findings suggest that this method could be used to determine the level of monitoring that is needed for any patient admitted for consideration of AWS.
A hallmark of AWS is autonomic instability. Therefore, the finding that cg07375256 maps to the transcription start site region of ZSCAN25 (previously known as ZNF498), a gene encoding a zinc finger protein previously associated with hypertension, fits well to our clinical understanding of AWS [Citation44]. Still, it is important to realize that associations are not causations, and that even in our examination of entry and exit methylation in 45 pairs of DNA samples, methylation at 18 other loci also significantly reverted during the three-week period of hospitalization-induced abstinence. Furthermore, the methylation changes observed were in WBCs, not the neuronal or vascular tissue mediating the autonomic instability or occurrence of seizures. Therefore, a truly thorough understanding of the relationship of changes of DNA methylation to the more global clinical signs and symptoms of AWS will likely require more in-depth integrated human/animal model studies. Still, these data demonstrating that Dcg07375256 methylation can predict serious AWS course (seizures, phenobarbital administration) are highly suggestive that these efforts would be fruitful.
The substantial reversion of methylation at cg07375256 within a three-week period (see ) in the DNA samples from our 2014 and 2019 studies [Citation9,Citation27] (see and ) may suggest the reason why other biomarkers have failed or perform more poorly in predicting serious AWS. Both CDT and Peth have relatively short half-lives, on the order of 7–10 days, and 4–10 days, respectively [Citation21,Citation25]. Although the earlier pioneering works of the Isbell and Mendelson groups are somewhat limited, they do suggest that long periods of sustained heavy drinking, on the order of 15 drinks per day for more than 3 weeks is necessary to become vulnerable to serious AWS. If so, the short window of time recognized by the CDT and PEth assays may be insufficient to capture the critical period of usage. At the other extreme, the markers in the ATS, were calibrated to predict chronic HAC, which was defined as drinking at least eight drinks per day for at least 6–8 weeks. As a consequence of this selection for more sustained drinking, the markers in the ATS panel all have reversion half-lives of 3 months or more [Citation9]. But in this study, the ATS only modestly predicts seizures and does not significantly predict the likelihood of the patient receiving phenobarbital. This suggests that the longer half-life markers are also unsuited for ideally predicting AWS. The methylation half-life of cg07375256, which appears to be approximately 3–4 weeks based off these and our 2014 studies, may be at the happy medium and thereby particularly well suited to capture heavy sustain alcohol consumption during the critical time window in which induction of vulnerability to severe AWS takes place. Further studies to better understand the reversion of alcohol-induced DNA methylation changes in response to alcohol abstinence and the biology of AWS could shed additional insight.
Still, it is important to realize that while Dcg07375256 methylation seems to predict severe AWS outcomes such as seizures or the choice of clinicians to use phenobarbital, there may be other methylation markers or combination of methylation markers that predict better. For example, we choose to develop an MSdPCR marker for cg07375256 instead of cg09807356, based on our experience as clinical translational scientists. It may well be that an MSdPCR assay targeting cg09807356 or a combination of the two assays or other clinical variables might predict outcomes even better. Furthermore, seizures and use of phenobarbital are only two of several types of indicators of severe AWS outcome. It may be that a broader predictor set may help to foretell other severe outcomes, such as delirium tremens, better.
Although it is likely that the level of alcohol needed to induce vulnerability to DTs or other severe outcomes may be similar to that for seizures, there may be other co-factors, such as the absence of key vitamins or genetic variation, that influence the likelihood of severe AWS outcomes such as seizures and DTs differently. Determining the presence of these additional factors may be a challenging endeavour. We note that the presence of hallucinations alone was not associated with any other clinical factor examined, suggesting routine clinical assessment of this symptom alone may be of limited utility, perhaps owing to its subjective nature.
At first glance, the finding that PAWSS values were associated with total benzodiazepine dose and BAC levels is intriguing. However, the PAWSS questionnaire specifically asks whether patients have used downers, including benzodiazepines, and have a BAC level of greater than 200 mg/dl. As such, the positive association of the PAWSS with these two variables may just reflect part-whole correlations, or alternatively, the tendency of patients who have received benzodiazepines in the past to manifest behaviours so that they receive them in the current hospitalization. Nevertheless, the failure of the PAWSS to predict any severe outcomes in this study suggests that its clinical utility may be limited.
A striking finding is the lack of association of seizures, the use of phenobarbital and any of the four biological markers of alcohol consumption (BAC, the ATS, the CDT, and Dcg07375256), with subject reports of alcohol consumption. Although other interpretations are possible, the most parsimonious explanation is that the subject reports of their alcohol consumption are not reliable. Indeed, in our clinical experience (BP, AA, and RP), we have found that patients often misrepresent their alcohol use history. Although others may disagree, in our research experience, we standardly employ multiple biomarkers of alcohol consumption including the CDT and epigenetic biomarkers to assess alcohol use in our subjects and routinely find much stronger relationships between biomarkers of alcohol use to one another and to key outcomes than to those found with self-report [Citation10,Citation12,Citation45]. When considering the circumstances in which most of the subjects in this study were presented, this is certainly understandable. Specifically, originally, many of the subjects did not willingly present for treatment and may fear the information being used against them, they may be reluctant to fully disclose their past usage of alcohol. Or even more understandably, the subjects in this study simply may not remember how much they drank. This should not be surprising. For example, even under the best of circumstances, dietary reports by sober patients have high rates of error [Citation46,Citation47]. Given the level of chronic intoxication that many of these subjects report, our subjects simply may not recall how much they have been drinking or how long they have been drinking. In any case, it is clear that in this study the biological variables (CDT, ATS, and Dcg07375256) tend to be positively associated with one another, while self-report of alcohol use only correlates with, unsurprisingly, scales such as the PAWSS that are based in part on self-report.
A strength of this study is the use of MSdPCR to assess cg07375256 status. A substantial barrier impeding the use of PEth as a biomarker is the need for mass spectroscopy for the most accurate determinations [Citation18,Citation22]. As a result, PEth determinations are done only in specialized testing settings thus making the likelihood of receiving test results in clinically meaningful time frame moot. Similarly, the CDT levels used in this study were determined using capillary electrophoresis [Citation39]. Although the machinery for these determinations appears to be more widespread than those for PEth, none of the local hospitals that we surveyed directly offered the testing service and instead relied on third party national testing companies, such as LabCorp (https://www.labcorp.com/), to provide the assessments. In contrast, MSdPCR can be done in a matter of hours by any laboratory with access to a digital PCR system. In prior years, these dPCR systems were uncommon. However, over the past year, five manufacturers have entered the dPCR market with the list price of the most affordable systems being under $90,000. As such, there have been a growing number of FDA approved and Laboratory Developed Tests (LDT) designed for these systems.
Limitations of this study include that it is a naturalistic approach of the care by dozens of clinicians that required individual review of patients’ charts by the study team. As a result, it is possible that variation in provider behaviour could have affected study outcomes. It is also a single-site study of largely European American subjects from a relatively high socioeconomic status catchment area. Additionally, the subjects’ in-hospital clinicians were not members of the study team and thus some variability in charting of diagnoses and clinical outcomes was likely introduced. The number of seizure cases was also very small. Replication and extension in much larger dataset will be necessary to arrive at firm conclusions as to the robustness of the findings. In the case of more complex diagnostic criteria, such as those for delirium tremens (DTs), the lack of a firm operational case definition and variation in charting may also have weakened our ability to detect associations between cg07375256 methylation and clinical outcomes. Finally, and perhaps obviously, the use of BZDs and phenobarbital likely forestalled many severe events that would have occurred in the absence of treatment.
Obviously, the next steps in this line of endeavour are to enlarge both the size and diversity of the sample, while considering whether adding additional clinical information or biological predictors will aid prediction. Specifically, it may be that the addition of more methylation markers increases the power to predict severe AWS. Furthermore, whereas we are pleased that all subjects in our study survived to discharge, we note that even with good treatment, severe AWS can be fatal [Citation48]. Still, understanding whether this approach also predicts mortality will be critical.
In summary, we report that methylation status at cg07375256 predicts severe AWS outcomes and AWS-related seizures. Independent testing in larger, more diverse populations are in order.
Availability of data and materials
The data described herein are available upon reasonable request to Dr. Philibert and may be restricted to non-commercial uses.
Supplementary_Table_1_page.docx
Download MS Word (16.8 KB)Acknowledgments
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors would like to thank Drs. Steven R.H. Beach and Thad Abrams for their assistance in editing the manuscript.
Disclosure statement
Dr. Philibert is the Chief Executive Officer of Behavioral Diagnostics. The use of cg05575921 to assess smoking status is covered by existing and pending patents including US Patents 8,637,652 and 9,273,358. Similarly, the use of DNA methylation to assess alcohol is covered by existing and pending patents including European Union Patent 3,149,206. On behalf of Drs. Philibert and Andersen, Behavioral Diagnostics and the University of Iowa have filed an intellectual property claim on the use of DNA methylation to predict AWS and related phenomena.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592294.2023.2298057
Additional information
Funding
References
- Academic ED SBIRT Research Collaborative. The impact of screening, brief intervention, and referral for treatment on emergency department patients’ alcohol use. Ann Emerg Med. 2007; 50:699–710. e696. doi: 10.1016/j.annemergmed.2007.06.486
- Suen LW, Makam AN, Snyder HR, et al. National prevalence of alcohol and other substance use disorders among emergency department visits and hospitalizations: NHAMCS 2014–2018. J Gen Intern Med. 2022;37(10):2420–14. doi: 10.1007/s11606-021-07069-w
- Carlson RW, Kumar NN, Wong-Mckinstry E, et al. Alcohol withdrawal syndrome. Crit Care Clin. 2012;28(4):549–585. doi: 10.1016/j.ccc.2012.07.004
- Schuckit MA. Recognition and management of withdrawal delirium (delirium tremens). New England Journal of Medicine. 2014 Nov 27;371(22):2109–13.
- Kosten TR, O’Connor PG. Management of drug and alcohol withdrawal. N Engl J Med. 2003;348(18):1786–1795. doi: 10.1056/NEJMra020617
- Wood E, Albarqouni L, Tkachuk S, et al. Will this hospitalized patient develop severe alcohol withdrawal syndrome?: the rational clinical examination systematic review. JAMA. 2018;320(8):825–833. doi: 10.1001/jama.2018.10574
- Maldonado JR, Sher Y, Ashouri JF, et al. The “prediction of alcohol withdrawal severity scale”(PAWSS): systematic literature review and pilot study of a new scale for the prediction of complicated alcohol withdrawal syndrome. Alcohol. 2014;48(4):375–390. doi: 10.1016/j.alcohol.2014.01.004
- Wetterling T, Weber B, Depfenhart M, et al. DEVELOPMENT of a RATING SCALE to PREDICT the SEVERITY of ALCOHOL WITHDRAWAL SYNDROME. Alcohol Alcohol. 2006;41(6):611–615. doi: 10.1093/alcalc/agl068
- Philibert R, Miller S, Noel A, et al. A four marker digital PCR toolkit for detecting heavy alcohol consumption and the effectiveness of its treatment. J Insur Med. 2019;48(1):90–102. doi: 10.17849/insm-48-1-1-1.1
- Miller S, Mills JA, Long J, et al. A comparison of the predictive power of DNA methylation with carbohydrate deficient transferrin for heavy alcohol consumption. Epigenetics. 2020;16(9):969–979. doi: 10.1080/15592294.2020.1834918
- Lei M-K, Gibbons FX, Gerrard M, et al. Digital methylation assessments of alcohol and cigarette consumption account for common variance in accelerated epigenetic ageing. Epigenetics. 2022;17(13):1–15. doi: 10.1080/15592294.2022.2100684
- Beach SR, Ong ML, Gibbons FX, et al. Epigenetic and proteomic biomarkers of elevated alcohol use predict epigenetic aging and cell-type variation better than self-report. Genes. 2022;13(10):1888. doi: 10.3390/genes13101888
- Isbell H, Fraser HF, Wikler A, et al. An experimental study of the etiology of “rum fits” and delirium tremens. Q J Stud Alcohol. 1955;16(1):1–33. doi: 10.15288/qjsa.1955.16.001
- Mendelson JH, La Dou J. Part 1. Background and experimental design. J Stud Alcohol Suppl. 1964;25(S2):1–13. doi: 10.15288/qjsas.1964.s2.001
- Turner RC, Lichstein PR, Peden JG, et al. Alcohol withdrawal syndromes. J Gen Intern Med. 1989;4(5):432–444. doi: 10.1007/BF02599697
- Grüner Nielsen D, Andersen K, Søgaard Nielsen A, et al. Consistency between self-reported alcohol consumption and biological markers among patients with alcohol use disorder – a systematic review. Neuroscience & Biobehavioral Reviews. 2021;124:370–385. doi:10.1016/j.neubiorev.2021.02.006
- Verheij C, Haagsma JA, Koch BCP, et al. Screening for hazardous alcohol use in the emergency department: comparison of phosphatidylethanol with the alcohol use disorders identification test and the timeline follow-back. Alcoholism Clin & Exp Res. 2022;46(12):2225–2235. doi: 10.1111/acer.14958
- Cabarcos P, Álvarez I, Tabernero MJ, et al. Determination of direct alcohol markers: a review. Anal Bioanaly Chem. 2015;407(17):4907–4925. doi: 10.1007/s00216-015-8701-7
- Niemelä O, Alatalo P. Biomarkers of alcohol consumption and related liver disease. Scand J Clin Lab Invest. 2010;70(5):305–312. doi: 10.3109/00365513.2010.486442
- Novak L, Soravia LM, Bünter A, et al. Alcohol biomarker Phosphatidylethanol as a predictor of the severity of alcohol withdrawal syndrome. Alcohol Alcohol. 2023;58(2):198–202. doi: 10.1093/alcalc/agac071
- Helander A, Böttcher M, Dahmen N, et al. Elimination characteristics of the alcohol biomarker phosphatidylethanol (PEth) in blood during alcohol detoxification. Alcohol Alcohol. 2019;54(3):251–257. doi: 10.1093/alcalc/agz027
- Helander A, Zheng Y. Molecular species of the alcohol biomarker phosphatidylethanol in human blood measured by LC-MS. Clin Chem. 2009;55(7):1395–1405. doi: 10.1373/clinchem.2008.120923
- Karagülle D, Heberlein A, Wilhelm J, et al. Biological markers for alcohol withdrawal seizures: a retrospective analysis. Eur Addict Res. 2012;18(3):97–102. doi: 10.1159/000335273
- Wetterling T, Kanitz R-D, Renner F, et al. Does carbohydrate-deficient transferrin predict the severity of alcohol withdrawal syndrome? Alcoholism Clin & Exp Res. 1998;22(5):1053–1056. doi: 10.1111/j.1530-0277.1998.tb03698.x
- Moon H-W, Yun Y-M, Kim S, et al. Determination of carbohydrate-deficient transferrin levels by using capillary electrophoresis in a Korean population. Korean J Lab Med. 2010;30(5):477–484. doi: 10.3343/kjlm.2010.30.5.477
- Philibert R, Dogan M, Noel A, et al. Genome-wide and digital polymerase chain reaction epigenetic assessments of alcohol consumption. Am J Med Genet B Neuropsychiatr Genet. 2018;177(5):479–488. doi: 10.1002/ajmg.b.32636
- Philibert R, Penaluna B, White T, et al. A pilot examination of the genome-wide DNA methylation signatures of subjects entering and exiting short-term alcohol dependence treatment programs. Epigenetics. 2014;9(9):1212–1219. doi: 10.4161/epi.32252
- Pidsley R, Y Wong CC, Volta M, et al. A data-driven approach to preprocessing illumina 450K methylation array data. BMC Genomics. 2013;14(1):1–10. doi: 10.1186/1471-2164-14-293
- Triche JRT. FDb.InfiniumMethylation.hg19: annotation package for illumina infinium DNA methylation probes. R package version 2.2.0. 2014 [cited 2022 Jul 1]. Available from: https://bioconductor.org/packages/release/data/annotation/html/FDb.InfiniumMethylation.hg19.html
- Davis S, Du P, Bilke S, et al. methylumi: Handle Illumina methylation data. R package version 2.22.0. 2017 [cited 2022 Jul 1]. Available from: https://bioconductor.statistik.tu-dortmund.de/packages/3.5/bioc/html/methylumi.html
- Dogan M, Beach S, Simons R, et al. Blood-based biomarkers for predicting the risk for five-year incident coronary heart disease in the framingham heart study via machine learning. Genes. 2018;9(12):641. doi: 10.3390/genes9120641
- Dogan MV, Grumbach IM, Michaelson JJ, et al. Integrated genetic and epigenetic prediction of coronary heart disease in the framingham heart study. PLoS One. 2018;13(1):e0190549. doi: 10.1371/journal.pone.0190549
- Bonferroni C. Teoria statistica delle classi e calcolo delle probabilita. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commericiali di Firenze. 1936;8:3–62.
- Student, S. The probable error of a mean. Biometrika. 1908;6(1):1–25. doi: 10.2307/2331554
- Hamilton CM, Strader LC, Pratt JG, et al. The PhenX toolkit: get the most from your measures. Am J Epidemiol. 2011;174(3):253–260. doi: 10.1093/aje/kwr193
- Salzman C, Balter M, Ellinwood E, et al. American Psychiatric Association task force on benzodiazepine dependency, toxicity, and abuse. Washington (DC): American Psychiatric Press; 1990.
- Philibert R, Dawes K, Philibert W, et al. Alcohol use intensity decreases in response to successful smoking cessation therapy. Genes. 2021;13(1):2. doi: 10.3390/genes13010002
- Fang F, Andersen AM, Philibert R, et al. Epigenetic biomarkers for smoking cessation. Addiction Neuroscience. 2023;6:100079. doi:10.1016/j.addicn.2023.100079
- Schellenberg F, Wielders JP. Evaluation of capillary electrophoresis assay for CDT on SEBIA’s capillarys system: intra and inter laboratory precision, reference interval and cut-off. Clinica Chimica Acta. 2010;411:1888–1893. doi:10.1016/j.cca.2010.07.015 23–24
- Team RC. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. (ISBN 3-900051-07-0, 2014).
- Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12(1):77. doi: 10.1186/1471-2105-12-77
- Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102
- Oks M, Cleven KL, Healy L, et al. The safety and utility of phenobarbital use for the treatment of severe alcohol withdrawal syndrome in the medical intensive care unit. J Intensive Care Med. 2020;35(9):844–850. doi: 10.1177/0885066618783947
- Puck JM, Deschenes SM, Porter JC, et al. The interleukin-2 receptor γ chain maps to Xq13.1 and is mutated in X-linked severe combined immunodeficiency, SCIDX1. Hum Mol Genet. 1993;2(8):1099–1104. doi: 10.1093/hmg/2.8.1099
- Lei M-K, Gibbons FX, Gerrard M, et al. Digital methylation assessments of alcohol and cigarette consumption account for common variance in accelerated epigenetic ageing. Epigenetics. 2022;17(13):1991–2005. doi: 10.1080/15592294.2022.2100684
- St George SM, Van Horn ML, Lawman HG, et al. Reliability of 24-hour dietary recalls as a measure of diet in African-American youth. J Acad Nutr Diet. 2016;116:1551–1559. doi:10.1016/j.jand.2016.05.011
- Ravelli MN, Schoeller DA. Traditional self-reported dietary instruments are prone to inaccuracies and new approaches are needed. Front Nutr. 2020;7. doi: 10.3389/fnut.2020.00090
- Mainerova B, Prasko J, Latalova K, et al. Alcohol withdrawal delirium-diagnosis, course and treatment. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2015;159(1):044–052. doi: 10.5507/bp.2013.089
