ABSTRACT
Penile squamous cell carcinoma (SCC) is a rare and aggressive tumour mainly related to lifestyle behaviour and human papillomavirus (HPV) infection. Environmentally induced loss of imprinting (LOI) at the H19 differentially methylated region (H19DMR) is associated with many cancers in the early events of tumorigenesis and may be involved in the pathogenesis of penile SCC. We sought to evaluate the DNA methylation pattern at H19DMR and its association with HPV infection in men with penile SCC by bisulfite sequencing (bis-seq). We observed an average methylation of 32.2% ± 11.6% at the H19DMR of penile SCC and did not observe an association between the p16INK4a+ (p = 0.59) and high-risk HPV+ (p = 0.338) markers with methylation level. The average methylation did not change according to HPV positive for p16INK4a+ or hrHPV+ (35.4% ± 10%) and negative for both markers (32.4% ± 10.1%) groups. As the region analysed has a binding site for the CTCF protein, the hypomethylation at the surrounding CpG sites might alter its insulator function. In addition, there was a positive correlation between intense polymorphonuclear cell infiltration and hypomethylation at H19DMR (p = 0.035). Here, we report that hypomethylation at H19DMR in penile SCC might contribute to tumour progression and aggressiveness regardless of HPV infection.
Introduction
Penile squamous cell carcinoma (SCC) is a rare and aggressive neoplasia with increasing incidence in developing countries. Lifestyle conditions, such as poor hygiene, promiscuous sexual behaviour, and the presence of phimosis are the main risk factors associated with penile cancer [Citation1]. Human papillomavirus (HPV) infection is often related to penile SCC with a variable incidence in the male population from 11% to 87% which is slightly related to the diagnostic method [Citation2]. We recently reported that the incidence of HPV infection in the penile SCC population was 53.2% using a hybridization assay to capture high-risk HPV (hrHPV) and 22.3% using the p16INK4a marker, which does not affect the prognosis and survival rate [Citation3].
Owing to the sporadic aetiology of penile cancer, with an important environmental contribution, epigenetic alterations in gene expression control may trigger tumour development and tighter with genetic mutations propagate carcinogenesis and contribute to its aggressiveness [Citation4]. The biallelic expression of imprinted genes or loss of imprinting (LOI) is one of the most affected epigenetic processes that appear during early tumour development [Citation5]. The imprinting control regions (ICRs) are differentially methylated regions (DMRs) in a parent-of-origin manner leading to a monoallelic expression of the clustered imprinted genes [Citation6]. Since imprinted genes regulate cell differentiation [Citation7], metabolism [Citation8], proliferation [Citation9], and other biological processes, epigenetic alterations at the ICRs change the cell landscape and trigger carcinogenesis, being reported in many cancers such as lung [Citation10], colorectal [Citation11], glioblastoma [Citation12], and acute myeloid leukaemia [Citation13].
One of the well-recognized ICRs involved in carcinogenic transformation is the H19DMR (ICR1) [Citation14], mapped to human chromosome 11p15.5 region that controls the monoallelic expression of the clustered H19 and IGF2 genes [Citation15]. This region is localized at 2 kilobases upstream of the H19, a long non-coding RNA (lncRNA) with a controversial role during carcinogenesis, acting either as a tumour suppressor or oncogene [Citation16,Citation17]. The H19DMR contains multiple CTCF-binding domains (CCCTC-binding factor) with insulator activity and represents an enhancer competition model for gene regulation [Citation18]. The H19DMR is unmethylated at the maternal allele allowing the CTCF protein to bind to its DNA domains and H19 activation through enhancers located upstream of this gene. In the paternal allele, methylation promotes conformational changes near to H19 promoter region, which is silenced, while IFG2 is transcriptionally activated by the shared enhancers [Citation19].
In addition to its complex structural region, the H19DMR has two non-CpG single nucleotide polymorphisms (SNPs), the rs2107425:C>T and rs2071094:G>T, that are associated with parental allele-specific DNA methylation (ASM) status, thereby altering gene expression and chromatin remodelling [Citation20,Citation21]. However, ASM can be altered through environmental exposure and a high confluence of ASM regions has been reported, possibly due to a genetic variation at a regulatory SNP locus [Citation22]. The genomic variation in DMRs can be valuable as prognostic markers and may contribute to changes in DNA methylation pattern during carcinogenesis [Citation22]. Analysis based on DNA methylation and SNPs on conjoint has guided the stratification into high and low-risk groups for breast cancer, supporting the relevant prognostic value of genomic and epigenomic combined analysis for cancer biomarkers [Citation23].
Considering the importance of the H19DMR in growth-related pathways and cell differentiation [Citation24], LOI or biallelic expression of imprinted genes has been associated with carcinogenic transformation [Citation25], tumour progression [Citation26], metastasis [Citation26], and resistance to treatment [Citation27]. We previously reported an increase in 5-methylcytosine (5mC) and a decrease in 5-hydroxymethylcytosine (5hmC), markers of global methylation and demethylation respectively, were previously observed in our penile SCC cohort. However, HPV infection seems not to affect 5mC or 5hmC epigenetic markers [Citation3]. These global changes may affect the methylation pattern at ICRs in penile SCC. Thus, we sought to evaluate the DNA methylation at H19DMR and its association with clinical aspects of penile SCC and HPV infection.
Materials and methods
Study design and data collection
This is a retrospective study that included 30 penile SCC samples from patients who underwent partial or total penectomy, enlarged prostatectomy, or emasculation due to penile SCC, without any prior history of chemotherapy or radiotherapy and with detailed clinicopathological and follow-up data available, from 2015 to 2018 at Hospital Haroldo Juaçaba, Ceará, Brazil. The samples used came from the Hospital tumour tissue sample bank, stored at −80°C. The study was approved by the ethics committee of the Federal University of Ceará and Haroldo Juaçaba Hospital, according to process number 2.427.846. Data from the anatomopathological reports and medical records were collected for epidemiological evaluation. Pathological staging was performed according to the eighth edition of the American Joint Committee on Cancer (AJCC).
p16INK4a expression and high-risk HPV identification assays
The immunohistochemistry (IHC) assay was performed to p16INK4a protein expression using the anti-p16 antibody clone E6H4 (Roche CINtec® Histology) according to the manufacturer’s instructions. The p16INK4a expression must be ≥ 75% to be considered positive, with continuous and complete cytoplasmic and nuclear staining [Citation28]. Chromogenic in situ hybridization (CISH) was used to identify high-risk HPV (hrHPV) using the Ventana Inform HPV III Family 16 Probe diagnostic kit (Ventana Medical Systems, Tucson, AZ) (genotypes 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58 and 66). High-grade cervical intraepithelial neoplasia was used as a positive control for hrHPV. Skeletal striated muscle was used as a negative control for both assays. All the experiments were analysed by a blinded observer.
Isolation of genomic DNA
Genomic DNA from penile SCC samples was isolated using the salting-out protocol. Briefly, samples were lysed (80 mL proteinase-K buffer [0.375 M NaCl, 0.12 M EDTA, pH 8.0], 8 mL proteinase K [25 mg/mL], 10 mL 20% sodium dodecyl sulphate, and 280 mL H2O) overnight at 55°C with shaking. The samples were cooled down and 120 mL of 5 M NaCl was added. The samples were then shaken vigorously for 8 seconds and centrifuged (13,000 g for 5 minutes at 4°C). Next, 400 mL of the supernatant was mixed with 1 mL of 99% cold ethyl alcohol, inverted a few times, and kept at 20°C overnight. The precipitated DNA was washed twice with 70% cold ethyl alcohol and centrifuged (13,000 g for 5 minutes at 4°C). The concentration and quality of the isolated DNA were evaluated using a spectrophotometer (Nanodrop 2000c, Thermo Fisher Scientific, USA). The genomic DNA was stored at −20°C until they were used.
Bisulfite conversion and PCR amplification
Genomic DNA was used for DNA bisulfite conversion using the EZ DNA Methylation-Lightning kit (Zymo Research, USA) according to the manufacturer’s instructions. The amplification of the H19DMR was performed using Platinum Taq DNA polymerase (Invitrogen, USA) with primers containing Nextera (Illumina, USA) adapters (Supplementary Table S1) as previously described elsewhere [Citation29]. The PCR conditions to amplify the H19DMR were 95°C for 5 minutes, 50 cycles of denaturation at 94°C for 45 seconds, annealing at 59°C for 45 seconds, and extension at 72°C for 45 seconds, and a final extension step at 72°C for 10 minutes. The amplification of H19DMR was confirmed by electrophoresis on 1.5% agarose gel.
Next-generation sequencing
Targeted bisulfite sequencing was performed using the MiSeq platform (Illumina, San Diego, CA, USA), covering 23 CpG sites at H19DMR (chr11:1,999,757–2,000,060). PCR products, containing the adapters, were barcoded using the Illumina Nextera XT library preparation kit (Illumina, USA), and the sequencing was performed using the 600bp V3 reagents kit according to the manufacturer’s instructions. The FASTQ files for individual samples were generated using Illumina’s pipeline (bcl2fastq2-v2–20). The adapter and indexes were removed from the sequence using Trimmomatic v0.38.1 [Citation30]. The paired read sequences were merged using the default settings of FLASH v1.2.11.4 and aligned to the bisulfite converted genome using Bismark v0.18.2 with the following settings: –ambig_bam, which was also used to count the reads with different methylation percentage. Reads were considered methylated with ≥ 61% methylated CpG sites, partially methylated between 31% − 60% methylated CpG sites, and unmethylated with ≤ 30% methylated CpG sites. Visualization of the methylated CpGs in the regions of interest was performed based on Tabsat v1.0.2. The two single nucleotide polymorphism (SNP) genotyping, rs2107425 (C>T; Chr11:1999845) and rs2071094 (G>T; Chr11:1999934), were visualized with Integrative Genomics Viewer (IGV). For methylation analyses, positive (+) samples for either p16INK4a+ or hrHPV+ were mentioned as HPV+. Negative samples (-) for both marks were considered HPV-.
CTCF consensus binding sites search and in silico analysis of gene expression
CTCF binding sites in H19DMR were detected using the scan settings of the CTCFBSDB 2.0 database (https://insulatordb.uthsc.edu/) [Citation31], a comprehensive collection of experimentally determined and computationally predicted CTCF binding sites from the literature. The database uses sex position weight matrices (PWM) to report the single best hit in the query sequence. Usually, a short sequence with a PWM score > 3.0 is a suggestive match. Query sequence and database output result are available in Supplementary Table S2. In silico analyses of CTCF, H19, and IGF2 expression were performed in squamous cell carcinoma as per available data of RNA-seq from the OncoDB database (https://oncodb.org/) [Citation32]. Expression of three tumour types and their respective non-tumour tissues were accessed (Supplementary Table S3): head and neck squamous cell carcinoma (HNSC) (520 tumour samples and 44 non-tumour samples), cervical squamous cell carcinoma (CESC) (304 tumour samples and 22 non-tumour samples), and lung squamous cell carcinoma (LUSC) (503 tumour samples and 51 non-tumour samples). Expression data were normalized through transcripts per million (TPM).
Statistical analysis
Student’s t-test was used to compare the average number of methylated, unmethylated, and partially methylated reads between HPV positive and negative groups and in silico gene expression. Fisher’s exact test was applied to verify the association of clinical variables and the SNPs rs2107425 and rs2071094 with methylation pattern at H19DMR. The Kaplan – Meier curves were predicted by the log-rank (Mantel-Cox) test and linear regression was used to correlate H19DMR methylation levels and age. Statistical analyses were carried out using GraphPad Prism 8.4.2 (Intuitive Software for Science, San Diego, California, USA). p < 0.05 was considered statistically significant.
Results
The clinical characteristics, pathological classification, and HPV infection of penile SCC participants are presented in . The average age of patients was 63.8 (± 18.8) years old and the most frequent primary tumour staging grades were pT2 (40%; 12/30) and pT3 (36.7%; 11/30). Lymph node metastasis was observed in 51.9% (14/27) of the participants, followed by locoregional recurrence (29.7%; 7/27) and systemic metastasis (15.4%; 4/26). Immune cell infiltration was mostly mild and moderate for peritumoral lymphocyte infiltrate (PLI) (60%; 18/30) and intratumoral polymorphonuclear infiltrate (IPI) (53.3%; 16/30). HPV infection was observed in 40% (12/30) of the participants using the p16INK4a+ marker and 53.2% (16/30) using hrHPV+ and 63.3% (19/30) were positive for at least one of the tests.
Table 1. Clinical and pathological aspects of penile squamous cell carcinoma participants.
The average methylation was 32.2% ± 11.6% in all samples, however, 43.3% of penile SCC showed methylation levels lower than 30%. The mean number of unmethylated reads was higher, representing 63.3% ± 10.6% of all reads, and the mean number of partially methylated reads was 4.5% ± 5.8 (). Methylation level at H19DMR was not associated with HPV infection using both p16INK4a + (p = 0.59) and hrHPV (p = 0.338) markers. However, reduced H19DMR methylation level was positively correlated with intense intratumoral polymorphonuclear infiltrate (IPI) (p = 0.035), but with no other clinical and pathological variables (). When the samples were stratified into those below and above 60 years of age, no correlation was observed between the methylation levels of 30% and 60% (). Considering all ages, a negative correlation between DNA methylation and age was observed (Supplementary Figure S1). The average distribution of methylated (p = 0.15), unmethylated (p = 0.38), and partially methylated reads (p = 0.06) was not different between HPV+ (p16INK4a+ or hrHPV+) and HPV- samples () and the methylation average did not change according to HPV positive (35.4% ± 10%) and negative (32.4% ± 10.1%) groups ().
Figure 1. Bisulfite sequencing of H19DMR in penile squamous cell carcinoma. Each row represents a sample (S) and its HPV status (positive samples were considered as p16INK4a+ or hrHPV+). The number of reads is specified for each sample. The SNPs position are specified by the red arrow.
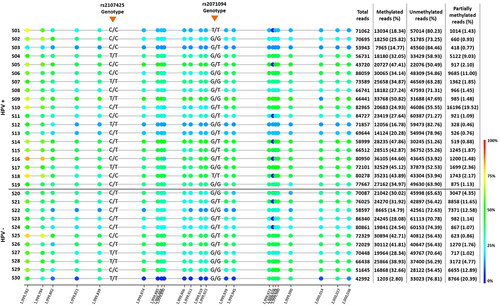
Figure 2. Average of methylated, unmethylated and partially methylated reads in HPV positive (HPV+) and negative (HPV-) of penile squamous cell carcinoma. Positive samples were considered as p16INK4a+ or hrHPV+.
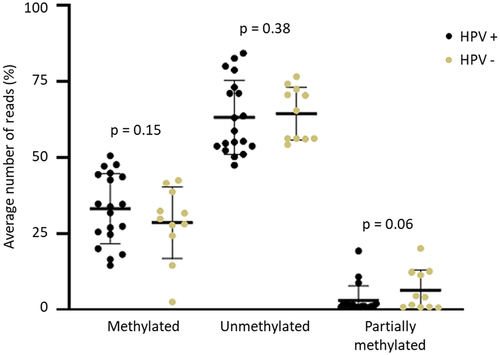
Figure 3. Percentage of methylation for each CpG site in HPV positive (HPV+) and negative (HPV-) of penile squamous cell carcinoma. The CTCF binding site is indicated by the grey arrow. Positive samples were considered as p16INK4a+ or hrHPV+.
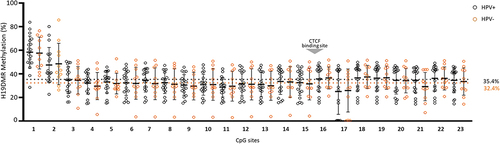
Table 2. Correlation between clinical data of penile squamous cell carcinoma participants and methylation level at H19DMR.
Reduced methylation level (hypomethylation) was observed for most CpG sites in the H19DMR, however, the CpG1 (Chr11: 1,999,782), CpG2 (Chr11: 1,999,794), and CpG17 (Chr11: 1,999,976) showed a different pattern in both HPV+ (CpG1 58.4% ± 13.6%, CpG2 47.4% ± 14.9%, and CpG17 25.1% ± 19.3%), and HPV- (CpG1 57.6% ± 13.4%, CpG2 48.4% ± 17.5%, and CpG17 25.9% ± 18.2%) (). Interestingly, the sequenced region presents a potential binding site for the CTCF factor (Chr11: 1,999,964–1,999,973), which comprises exactly the CpG16 site (Chr11: 1,999,973) (; Supplementary Table S2). Additionally, the genotypic frequency of both rs2107425 (C>T) and rs2071094 (G>T) SNPs was not related to changes in the methylation pattern at H19DMR (). Hypomethylation at H19DMR and SNPs genotype did not affect the survival rate in penile SCC ().
Figure 4. Kaplan – Meier curve for survival probability (n = 27). (a) methylation levels; (b) rs2107425 genotype; (c) s2071094 genotype.
Table 3. Correlation between SNPs genotype (rs2107425 and rs2071094) and H19DMR methylation pattern.
In silico analysis of gene expression in three different squamous cell carcinomas showed an increased expression of CTCF in HNSC (p = 1.7e-10) and LUSC (p = 1.4e-08) in comparison to non-tumour samples. H19 showed reduced expression in HNSC (p = 3.5e-02) and increased expression in LUSC (p = 8.5e-06) when compared to their respective non-tumour samples. IGF2 had lower expression in CESC (p = 1.2e-05) and increased expression in LUSC (p = 1.1e-02) when compared to non-tumour samples (Supplementary Table S3).
Discussion
LOI is associated with the early events of tumorigenesis due to the important role of imprinted genes in cell differentiation and growth. We reported a hypomethylated pattern at H19DMR in penile SCC, that was not related to HPV infection considering both p16INK4a and hrHPV markers. The hypomethylation was correlated to IPI but with no other clinical characteristics or survival rate. Additionally, the average methylation between the CpG sites was lower and similar, however, three specific sites showed a different pattern (CpG1, CpG2, and CpG17). The genotypic frequency of the SNPs rs2107425 (C>T) and rs2071094 (G>T) was not related to the methylation level at H19/IGF2 DMR.
The H19DMR gene cluster is related to the maintenance of cellular processes such as cell growth and proliferative activity [Citation33]. Changes in the methylation levels at this locus or LOI affect the monoallelic expression of both H19 and IGF2 genes [Citation34]. The impact of LOI at H19DMR has been previously linked to an increased IGF2 gene expression in prostate, colorectal, and rectum cancer [Citation35,Citation36]. In colorectal cancer, the increase in IGF2 protein expression via LOI is related to an increase in the carcinogenic pathways AKT1 (AKT serine/threonine kinase 1), IR-A (insulin receptor A), and WNT/beta-catenin. Given the modulatory role in such pathways, the IGF2 gene has been seen as a potential anticancer target. Xenographic models with increased IGF2 expression showed high inhibition of tumour growth and tumour regression when treated with new anti-IGF2 targets, such as BI 885,578, MEDI-573, and anti-VEGF therapy [Citation37,Citation38].
Loss of methylation at H19DMR has also been observed in bladder tumours. Byun et al. (2007) showed 20% of hypomethylation at the paternal allele when compared to normal mucosa in matched cases of bladder tumours [Citation14]. Biallelic expression of the lncRNA H19 or the inactivation of its maternally active copy due to LOI at H19DMR is the main alteration behind the preneoplastic Beckwith – Wiedemann syndrome [Citation39] and the paediatric Wilms’ tumour and rhabdomyosarcoma [Citation40]. The lncRNA H19 is a known oncofetal gene, with an intricate role during tumorigenesis, acting as a tumour suppressor in cancer initiation [Citation16] and as an oncogene during malignant progression [Citation41]. The biallelic expression of the lncRNA H19 due to LOI increases the risk of colorectal [Citation42], bladder [Citation43], breast and oral squamous cell carcinoma , and acute myeloid leukaemia (LMA), among others [Citation44]. Recently, we have shown that the knockdown of the lncRNA H19 gene and its reduced expression increases cell proliferation and metaphase translocation events [Citation33].
As the region analysed has a binding site for the CTCF protein, the hypomethylation at the surrounding CpG sites might alter its insulator function [Citation18]. This alteration may promote the biallelic expression of the lncRNA H19 and the silencing of the IGF2 in penile SCC. Furthermore, the hypomethylation observed in our current study seems to be independent of HPV infection, given the similar methylation levels between the HPV+ and HPV- samples considering either p16INK4a and hrHPV markers. Similarly, H19DMR hypomethylation was not associated with HPV infection in invasive cervical cancer (ICC) [Citation45] and cervical intraepithelial neoplasia (CIN) [Citation46]. We previously showed that the 5mC Global DNA methylation mark is increased in penile SCC and is not influenced by HPV infection [Citation3]. Although the genome-wide loss of CpG DNA methylation is an age-related event, our results did not show an association between age < 60 and >60 years old with methylation levels of 0% − 30% and 31% − 60%, but a negative correlation was observed between age and DNA methylation at H19DMR in penile SCC. Horvath (2013) reported that DNA methylation per tissue may differ from chronological age, and for squamous cell carcinomas, a lower time-dependent acceleration of methylation loss was observed when compared to other cancers [Citation47].
The H19DMR hypomethylation may influence the tumour microenvironment, as increased IPI was correlated with reduced methylation at this region. Likewise, immune cell infiltration in the tumour microenvironment was associated with DNA and RNA methylation modifications in colorectal cancer [Citation48] and gastric cancer [Citation49]. As this region controls the H19 and IFG2 imprinting cluster, LOI is related to altered glucose metabolism and diabetes [Citation50]. Intrauterine hyperinsulinemia changes H19DMR methylation in the foetuses which exhibited impaired glucose tolerance and insulin resistance [Citation51]. Obesity is a risk factor for cancer in general and is associated with an increased risk of invasive penile cancer, inducing chronic inflammation and insulin resistance [Citation52]. Overexpression of IGF1R was reported by Ball and colleagues (2016) in 62% of a cohort of 53 men diagnosed with penile cancer and was associated with inferior progression-free survival (PFS) [Citation53]. A comparison between patients without IGF1R overexpression and those with overexpression revealed a significant difference in 5-year PFS rates, with 94.1% versus 45.8%, respectively. In a subsequent study by the same research group, IGF1R overexpression was observed in approximately two-thirds of penile squamous cell carcinoma (SCC) cases among 112 patients. The findings indicated a noteworthy association with histologic subtype and grade, suggesting a worse prognosis for tumours exhibiting IGF1R overexpression [Citation54].
The HPV virus sequence can be integrated into the genome of the host cell, leading to mutational events, disrupted gene expression, and genomic instability [Citation55]. Furthermore, we also analysed two non-CpG SNPs (rs2107425 and rs2071094) at H19DMR previously reported as biomarkers for allele-specific DNA methylation (ASM) pattern [Citation56]. However, these SNPs were not related to ASM in penile SCC, as the genotype frequency was not different between HPV+ and HPV-, and it was not related to the methylated or unmethylated stretch. Canto et al. (2022) showed HPV-related mutations in penile cancer, and these alterations were localized to HPV integration sites (HPVis) and miRNA regions [Citation57]. A recent study showed that a high somatic tumour mutation burden (TMB) is associated with HPV-positive penile SCC, and these data show that the molecular scenario for this disease may depend on viral infection [Citation58]. The SNP rs1042522 (pArg72Pro) in the TP53 gene has been associated with cancer risk susceptibility, as the variant is more likely to be degraded via ubiquitinylation by the hrHPV E6 oncoprotein in various cancers [Citation59], but this association was not confirmed for penile SCC [Citation60].
The accumulation of myeloid-derived suppressor cells (MDSCs), such as polymorphonuclear neutrophils, is a hallmark of cancer, and changes in the tumour microenvironment associated with MDSC reduction have been reported in an in vitro model of penile SCC [Citation61,Citation62]. This marker has potent immunosuppressive effects, although the mechanisms that cause MDSC growth in the tumour microenvironment remain unknown. It has been reported that MDSCs respond to DNA methylation modulation, as the reduction of MDSC growth and accelerated activation of antigen-specific cytotoxic T cells are altered by the use of DNA methyltransferase inhibitors such as decitabine [Citation63]. Penile SCC DNA methylation signature should be further explored since the hypomethylation pattern at H19DMR in different types of tumours is associated with tumour progression, aggressiveness, and tumour microenvironment.
The identification of epigenetic reprogramming and molecular alterations in penile SCC, an extremely aggressive tumour with increased incidence in developing countries, whose therapies are limited and mostly based on surgical excision, can assist in disease diagnosis, prognosis, and treatment. To our knowledge, this is the first study that has evaluated the methylation pattern at H19DMR in penile SCC. However, some limitations need to be mentioned. First, we do not have the participant’s background information such as sociodemographic profile, sexual behaviour, HPV vaccination, and the identification of other related histological subtypes and tumour topography. Despite the high incidence of hrHPV in our cohort, two other viral genotypes, hrHPV 59 and 68, were not evaluated. Since we did not evaluate the expression of these imprinted genes in the samples (in vitro), we performed an in silico analysis in squamous cell carcinomas, similar to penile cancer, using the OncoDB database. H19 and IGF2 expression varied according to tumour origin as HNSC, LUSC, and CESC, while the insulator gene CTCF showed increased expression in both HNSC and LUSC. We could not find gene expression results in OncoDB in penile SCC, highlighting the importance of studies related to the molecular mechanisms of this tumour type. Despite the monoallelic pattern in most cell types, the expression of imprinted genes varies depending on the stage of development [Citation6]. The hypomethylation observed in this study may favour the expression of the H19 gene, an oncofetal lncRNA that is frequently overexpressed in many types of cancer, favouring carcinogenesis and tumour aggressiveness [Citation38,Citation39]. Given its relatively low incidence worldwide, but increased in underdeveloped countries, little is known about the genetic and epigenetic epidemiology of penile SCC, and future studies are needed to better understand the molecular profile of this tumour.
Ethics approval and consent to participate:
This study was approved by our Institutional Review Board (process number 2.427.846). The samples are kept in the custody of the Cancer Institute of Ceará Biobank, as well as the data from the medical records and the histopathological blocks. All patients agreed to the Informed Consent Form.
Financial support:
Programa Pesquisa para o SUS (PPSUS – process n°: 3931545/2017), Fundação Cearence de Apoio ao Desenvolvimento Científico e Tecnológico (FUNCAP, Process n°: UNI-0210–00054.01.00/23); Experimental Biology Center, University of Fortaleza; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Programa de Excelência Acadêmica (CAPES-PROEX) and Programa de Apoio à Pós-Graduação (CAPES-PROAP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Grant Numbers: 437037/2018–5 (C.L.M.F.) and 434,821/2018–7 (C.P.), besides the research sponsorships of C. P. (PQ-1B, Process n°: 303102/2013–6) and C.L.M.F (PQ-2 Process n°: 306289/2022–9).
Supplementary Table 1.docx
Download MS Word (21.4 KB)Supplementary Figure 1.tif
Download TIFF Image (64.9 KB)-)Supplementary Table 2.docx
Download MS Word (17.2 KB)-)Supplementary Table 3.docx
Download MS Word (17.3 KB)Acknowledgments
The authors are extremely grateful to the study participants and their families. Also, we would like to thank the members of the Laboratory of Genomics and Bioinformatics of the Experimental Biology Centre (NUBEX) at the University of Fortaleza (UNIFOR), the Experimental Oncology Laboratory (LOE), the Cancer Institute of Ceará (ICC), and the Genomics and Bioinformatics Centre (CeGenbio) of Drug Research and Development Centre (NPDM) at the Federal University of Ceará (UFC) for the support offered. We also would like to thank Silvana França dos Santos for laboratory support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data is available with the authors.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592294.2024.2305081.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Douglawi A, Masterson TA. Updates on the epidemiology and risk factors for penile cancer. Transl Androl Urol. 2017;6(5):785–12. doi: 10.21037/tau.2017.05.19
- Yanagawa N, Osakabe M, Hayashi M, et al. Detection of HPV-DNA, p53 alterations, and methylation in penile squamous cell carcinoma in Japanese men. Pathol Int. 2008;58(8):477–482. doi: 10.1111/j.1440-1827.2008.02259.x
- Santos S, Hirth CG, Pinheiro DP, et al. HPV infection and 5mC/5hmC epigenetic markers in penile squamous cell carcinoma: new insights into prognostics. Clin Epigenetics. 2022;1–12. [Internet]. BioMed Central. doi:10.1186/s13148-022-01360-1
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7(1):21–33. doi: 10.1038/nrg1748
- Lozano-Ureña A, Jiménez-Villalba E, Pinedo-Serrano A, et al. Aberrations of Genomic Imprinting in Glioblastoma Formation. Front Oncol. 2021;11:1–10. doi: 10.3389/fonc.2021.630482
- Monk D, Mackay DJG, Eggermann T, et al. Genomic imprinting disorders: lessons on how genome, epigenome, and environment interact. Nat Rev Genet. 2019;20:235–248. [Internet]. Springer US. doi: 10.1038/s41576-018-0092-0
- Sagi I, De Pinho JC, Zuccaro MV, et al. Distinct imprinting signatures and biased differentiation of human androgenetic and parthenogenetic embryonic stem cells. Cell Stem Cell. 2019;25(3):419–432.e9. doi: 10.1016/j.stem.2019.06.013
- Cleaton MAM, Edwards CA, Ferguson-Smith AC. Phenotypic outcomes of imprinted gene models in mice: elucidation of pre- and postnatal functions of imprinted genes. Annu Rev Genomics Hum Genet. 2014;15(1):93–126. doi: 10.1146/annurev-genom-091212-153441
- Uribe-Lewis S, Woodfine K, Stojic L, et al. Molecular mechanisms of genomic imprinting and clinical implications for cancer. Expert Rev Mol Med. 2011;13:1–22. doi: 10.1017/S1462399410001717
- Zhou J, Cheng T, Li X, et al. Epigenetic imprinting alterations as effective diagnostic biomarkers for early-stage lung cancer and small pulmonary nodules. Clin Epigenetics. 2021;13(1):1–14. doi: https://doi.org/10.1186/s13148-021-01203-5 [Internet]. BioMed Central
- Tian F, Tang Z, Song G, et al. Loss of imprinting of IGF2 correlates with hypomethylation of the H19 differentially methylated region in the tumor tissue of colorectal cancer patients. Mol Med Rep. 2012;5(6):1536–1540. doi: 10.3892/mmr.2012.833
- Zhu YF, Guo YB, Zhang HY, et al. Prognostic significance of contactin 3 expression and associated genes in glioblastoma multiforme. Oncol Lett. 2019;18:1863–1871. doi: 10.3892/ol.2019.10482
- Yang MY, Lin PM, Yang CH, et al. Loss of ZNF215 imprinting is associated with poor five-year survival in patients with cytogenetically abnormal-acute myeloid leukemia. Blood Cells Mol Dis. 2021;90:102577. doi: 10.1016/j.bcmd.2021.102577 [Internet]Elsevier Inc.
- Byun HM, Wong HL, Birnstein EA, et al. Examination of IGF2 and H19 loss of imprinting in bladder cancer. Cancer Res. 2007;67(22):10753–10758. doi: 10.1158/0008-5472.CAN-07-0329
- Nordin M, Bergman D, Halje M, et al. Epigenetic regulation of the Igf2/H19 gene cluster. Cell Prolif. 2014;47(3):189–199. doi: 10.1111/cpr.12106
- Yoshimizu T, Miroglio A, Ripoche MA, et al. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci U S A. 2008;105(34):12417–12422. doi: 10.1073/pnas.0801540105
- Tietze L, Kessler SM. The good, the bad, the question– H19 in hepatocellular carcinoma. Cancers (Basel). 2020;12(5):16. doi: 10.3390/cancers12051261
- Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet Nature Publishing Group. 2014;15(4):234–246. doi: 10.1038/nrg3663
- Yang Z, Zhang T, Han S, et al. Long noncoding RNA H19 – a new player in the pathogenesis of liver diseases. Transl Res. 2021;230:139–150. doi: 10.1016/j.trsl.2020.11.010
- Vohra M, Sharma AR, Prabhu BN, et al. SNPs in sites for DNA methylation, transcription factor binding, and miRNA targets leading to allele-specific gene expression and contributing to complex disease risk: a systematic review. Public Health Genomics. 2021;23(5–6):155–170. doi: 10.1159/000510253
- Zhong R, Liu L, Tian Y, et al. Genetic variant in SWI/SNF complexes influences hepatocellular carcinoma risk: a new clue for the contribution of chromatin remodeling in carcinogenesis. Sci Rep. 2014;4(1):1–6. doi: 10.1038/srep04147
- Do C, Dumont E, Salas M, et al. Allele-specific DNA methylation is increased in cancers and its dense mapping in normal plus neoplastic cells increases the yield of disease-associated regulatory SNPs. Genome Biol Genome Biology. 2020;21(1):1–39. doi: 10.1186/s13059-020-02059-3
- Shilpi A, Bi Y, Jung S, et al. Identification of genetic and epigenetic variants associated with breast cancer prognosis by integrative bioinformatics analysis. Cancer Inform. 2017;16:1–13. doi: 10.4137/CIN.S39783
- Yamaguchi Y, Tayama C, Tomikawa J, et al. Placenta-specific epimutation at H19-DMR among common pregnancy complications: its frequency and effect on the expression patterns of H19 and IGF2. Clin Epigenetics Clinical Epigenetics. 2019;11(1):1–13. doi: 10.1186/s13148-019-0712-3
- Wu J, Qin Y, Li B, et al. Hypomethylated and hypermethylated profiles of H19DMR are associated with the aberrant imprinting of IGF2 and H19 in human hepatocellular carcinoma. Genomics. 2008;91(5):443–450. doi: 10.1016/j.ygeno.2008.01.007
- Gao T, He B, Pan Y, et al. H19 DMR methylation correlates to the progression of esophageal squamous cell carcinoma through IGF2 imprinting pathway. Clin Transl Oncol. 2014;16(4):410–417. doi: 10.1007/s12094-013-1098-x
- Wang J, Ma X, Si H, et al. Role of long non-coding RNA H19 in therapy resistance of digestive system cancers. Mol Med. 2021;27:1–9. doi: 10.1186/s10020-020-00255-2 [Internet]. BioMed Central.
- Cubilla AL, Lloveras B, Alejo M, et al. Value of p16INK4a in the pathology of invasive penile squamous cell carcinomas: a report of 202 cases. Am J Surg Pathol. 2011;35(2):253–261. doi: 10.1097/PAS.0b013e318203cdba
- Vértesy Á, Arindrarto W, Roost MS, et al. Parental haplotype-specific single-cell transcriptomics reveal incomplete epigenetic reprogramming in human female germ cells. Nat Commun. 2018;9(1):1–10. doi: http://dx.doi.org/10.1038/s41467-018-04215-7 [Internet]. Springer US
- Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120.
- Ziebarth JD, Bhattacharya A, Cui Y. CTCFBSDB 2.0: a database for CTCF-binding sites and genome organization. Nucleic Acids Res. 2013;41(D1):D188–D194. doi: 10.1093/nar/gks1165
- Tang G, Cho M, Wang X. OncoDB: an interactive online database for analysis of gene expression and viral infection in cancer. Nucleic Acids Res. 2022;50(D1):D1334–9. doi: 10.1093/nar/gkab970 Oxford University Press.
- da Silva Santos R, Pascoalino Pinheiro D, Pinheiro Rodrigues Teixeira L, et al. CRISPR/Cas9 small promoter deletion in H19 lncRNA is associated with altered cell morphology and proliferation. Sci Rep. 2021;11(1):11. doi: https://doi.org/10.1038/s41598-021-97058-0 [Internet]. Nature Publishing Group UK.
- Bhusari S, Yang B, Kueck J, et al. Insulin-like growth factor-2 (IGF2) loss of imprinting marks a field defect within human prostates containing cancer. Prostate. 2011;185(4S):23. doi: 10.1016/j.juro.2011.02.1695
- Fu VX, Dobosy JR, Desotelle JA, et al. Aging and cancer-related loss of insulin-like growth factor 2 imprinting in the mouse and human prostate. Cancer Res. 2014;23:1–7.
- Belharazem D, Magdeburg J, Berton AK, et al. Carcinoma of the colon and rectum with deregulation of insulin-like growth factor 2 signaling: clinical and molecular implications. J Gastroenterol. 2016;51(10):971–984. doi: 10.1007/s00535-016-1181-5
- Haluska P, Menefee M, Plimack ER, et al. Phase I dose-escalation study of MEDI-573, a bispecific, antiligand monoclonal antibody against IGFI and IGFII, in patients with advanced solid tumors. Physiol Behav. 2015;20:4747–4757. Internet Carolina/Desktop/Artigos para acrescentar na qualificação/The impact of birth weight on cardiovascular disease risk in the.pdf: https://pubmed.ncbi.nlm.nih.gov/27165699%0Ahttps://www.ncbi.nlm.nih.gov/pmc/articles/PMC5664198/%0Afile:///C:/Users/Carla
- Sanderson MP, Hofmann MH, Garin-Chesa P, et al. The IGF1R/INSR inhibitor BI 885578 selectively inhibits growth of IGF2-overexpressing colorectal cancer tumors and potentiates the efficacy of anti-VEGF therapy. Mol Cancer Ther. 2017;16(10):2223–2233. doi: 10.1158/1535-7163.MCT-17-0336
- De Crescenzo A, Coppola F, Falco P, et al. A novel microdeletion in the IGF2/H19 imprinting centre region defines a recurrent mutation mechanism in familial Beckwith–Wiedemann syndrome. Eur J Med Genet. 2011;54(4):e451–4. doi: 10.1016/j.ejmg.2011.04.009 [Internet]. Elsevier Masson SAS
- Lynch CA, Tycko B, Bestor TH, et al. Reactivation of a silenced H19 gene in human rhabdomyosarcoma by demethylation of DNA but not by histone hyperacetylation. Mol Cancer. 2002;1(1):2–9. doi: 10.1186/1476-4598-1-2
- Matouk IJ, Raveh E, Abu-Lail R, et al. Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys Acta Mol Cell Res. 2014;1843(7):1414–1426. doi: 10.1016/j.bbamcr.2014.03.023 [Internet]. Elsevier B.V.
- Cui H, Onyango P, Brandenburg S, et al. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;2:6442–6446.
- Verhaegh GW, Verkleij L, Vermeulen SHHM, et al. Polymorphisms in the H19 Gene and the risk of bladder cancer. Eur Urol. 2008;54:1118–1126.
- Ghafouri-Fard S, Esmaeili M, Taheri M. H19 lncRNA : Roles in tumorigenesis. Biomed Pharmacother. 2020;123:123. doi: 10.1016/j.biopha.2019.109774
- Vidal AC, Henry NM, Murphy SK, et al. PEG1/MEST and IGF2 DNA methylation in CIN and in cervical cancer. Clin Transl Oncol. 2014;16(3):266–272. doi: 10.1007/s12094-013-1067-4
- Bosire C, Vidal AC, Smith JS, et al. Association between PEG3 DNA methylation and high-grade cervical intraepithelial neoplasia. Infect Agent Cancer. 2021;16(1):1–8. doi: 10.1186/s13027-021-00382-3
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):115. doi: 10.1186/gb-2013-14-10-r115
- Zou Q, Wang X, Ren D, et al. DNA methylation-based signature of CD8+ tumor-infiltrating lymphocytes enables evaluation of immune response and prognosis in colorectal cancer. J Immunother Cancer. 2021;9(9):1–13. doi: 10.1136/jitc-2021-002671
- Zhang B, Wu Q, Li B, et al. M6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer Molecular Cancer. 2020;19(1):1–21. doi: 10.1186/s12943-020-01170-0
- Wang B, Suen CW, Ma H, et al. The Roles of H19 in Regulating Inflammation and Aging. Front Immunol. 2020;11:1–11. doi: 10.3389/fimmu.2020.579687
- Jiang Y, Zhu H, Chen Z, et al. Hepatic IGF2/H19 epigenetic alteration induced glucose intolerance in gestational diabetes mellitus offspring via FoxO1 mediation. Front Endocrinol. 2022;13:1–11. doi: 10.3389/fendo.2022.844707
- Barnes KT, McDowell BD, Button A, et al. Obesity is associated with increased risk of invasive penile cancer. BMC Urol. 2016;16(1):7–10. doi: http://dx.doi.org/10.1186/s12894-016-0161-7 [Internet]. BMC Urology.
- Ball MW, Bezerra SM, Chaux A, et al. Overexpression of insulin-like growth factor-1 receptor is associated with penile cancer progression. Urology [Internet]. Elsevier Inc. 2016;92:51–56. doi: 10.1016/j.urology.2016.02.006
- Faraj SF, Gonzalez-Roibon N, Munari E, et al. Strong association of insulin-like growth factor 1 receptor expression with histologic grade, subtype, and HPV status in penile squamous cell carcinomas: a tissue microarray study of 112 cases. Virchows Arch Virchows Archiv. 2017;470(6):695–701. doi: 10.1007/s00428-017-2110-6
- Prati B, Marangoni B, Boccardo E. Human papillomavirus and genome instability: from productive infection to cancer. Clinics. 2018;73:1–9. doi: 10.6061/clinics/2018/e539s
- Shoemaker R, Deng J, Wang W, et al. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 2010;20(7):883–889. doi: 10.1101/gr.104695.109
- Canto Do LM, da Silva JM, Castelo-Branco PV, et al. Mutational signature and integrative genomic analysis of human papillomavirus-associated penile squamous cell carcinomas from Latin American patients. Cancers (Basel). 2022;14(14):14. doi: 10.3390/cancers14143514
- Nazha B, Zhuang T, Wu S, et al. Comprehensive genomic profiling of penile squamous cell carcinoma and the impact of human papillomavirus status on immune-checkpoint inhibitor-related biomarkers. Cancer. 2023;25(24):3884–3893. doi: 10.1002/cncr.34982 Springer US
- Khan Aroo MH, Khalil A, Rashid H. Evaluation of the p53 Arg72Pro polymorphism and its association with cancer risk: a HuGE review and meta-analysis. Genet Res (Camb). 2015;97:e7. doi: 10.1017/S0016672315000075
- Stoehr R, Weisser R, Wendler O, et al. P53 codon 72 polymorphism and risk for squamous cell carcinoma of the penis: a Caucasian case-control study. J Cancer. 2018;9(22):4234–4241. doi: 10.7150/jca.26050
- Ahmed ME, Falasiri S, Hajiran A, et al. Review the immune microenvironment in penile cancer and rationale for immunotherapy. J Clin Med. 2020;9(10):1–17. doi: 10.3390/jcm9103334
- Huang T, Cheng X, Chahoud J, et al. Effective combinatorial immunotherapy for penile squamous cell carcinoma. Nat Commun. 2020;11(1):11. doi: http://dx.doi.org/10.1038/s41467-020-15980-9 [Internet]. Springer US
- Smith AD, Lu C, Payne D, et al. Autocrine IL6-mediated activation of the STAT3–DNMT axis silences the TNFα–RIP1 necroptosis pathway to sustain survival and accumulation of myeloid-derived suppressor cells. Cancer Res. 2021;80(15):3145–3156. doi: 10.1158/0008-5472.CAN-19-3670
