ABSTRACT
Lung cancer is one familiar cancer that threatens the lives of humans. circCTNNB1 has been disclosed to have regulatory functions in some diseases. However, the functions and related regulatory mechanisms of circCTNNB1 in lung cancer remain largely indistinct. The mRNA and protein expression levels were examined through real-time polymerase chain reaction (RT-qPCR) and western blot. The cell proliferation was tested through CCK-8 assay. The cell migration and invasion were confirmed through Transwell assays. The cell senescence was evaluated through SA-β-gal assay. The binding ability between miR-186-5p and circCTNNB1 (or YY1) was verified through luciferase reporter and RIP assays. In this study, the higher expression of circCTNNB1 was discovered in lung cancer tissues and cell lines and resulted in poor prognosis. In addition, circCTNNB1 facilitated lung cancer cell proliferation, migration, invasion, and suppressed cell senescence. Knockdown of circCTNNB1 retarded the Wnt pathway. Mechanism-related experiments revealed that circCTNNB1 combined with miR-186-5p to target YY1. Through rescue assays, YY1 overexpression could rescue decreased cell proliferation, migration, invasion, increased cell senescence, and retarded Wnt pathway mediated by circCTNNB1 suppression. Furthermore, YY1 acts as a transcription factor that can transcriptionally activate circCTNNB1 to form YY1/circCTNNB1/miR-186-5p/YY1 positive loop. Through in vivo assays, circCTNNB1 accelerated tumour growth in vivo. All findings revealed that a positive loop YY1/circCTNNB1/miR-186-5p/YY1 aggravated lung cancer progression by modulating the Wnt pathway
Introduction
Lung cancer, as a familiar tumour, has the highest lethality [Citation1,Citation2]. In lung cancer, non-small cell lung cancer (NSCLC) takes up about 85% [Citation3]. In recent years, targeted therapies have been developed to lengthen the survival time of lung cancer patients, but the five-year survival rate has remained low [Citation4,Citation5]. Exploring the latent tumorigenic mechanisms for better bio-targets for lung cancer treatment is imperative.
Circular RNAs (circRNAs) are a kind of non-coding RNA, which is featured by covalently closed loops and participate in the tumorigenesis and development of cancers [Citation6,Citation7]. CircRNAs can be expressed, making them to be latent biomarkers for cancer progression, including lung cancer. circPOLA2 modulates miR-326/GNB1 axis to facilitate stemness in lung cancer [Citation8]. circBANP aggravates the progression of lung cancer through the miR-503/LARP1 axis [Citation9]. CircNEIL3 combines miR-1184 to enhance PIF1 expression, thereby regulating pyroptosis to affect radiotherapy in lung adenocarcinoma [Citation10]. Besides, circRNA C190 modulates the EGFR/ERK pathway to accelerate non-small cell lung cancer [Citation11]. CircCTNNB1 serves as a novel circRNA, and owns regulatory effects on some diseases. circCTNNB1 interacts with RBM15 to affect m6A modification, modulating aerobic glycolysis in osteosarcoma [Citation12]. Moreover, circCTNNB1 binds miR-96-5p to strengthen scavenger receptor class B type 1 (SRB1) expression, improving cerebral ischaemia/reperfusion injury [Citation13]. In spinal cord injury, circCTNNB1 modulates the Wnt/β-catenin pathway to affect neuronal injury [Citation14]. Furthermore, circCTNNB1 regulates DDX3-triggered transactivation of YY1 to exacerbate cancer progression [Citation15]. However, no report has investigated the role and related regulatory mechanisms of circCTNNB1 in lung cancer.
The miRNAs are another cluster of small non-coding RNAs (18∼22 nucleotides) that are important mediators regulated by lncRNAs or circRNAs to join in many biological processes in lung cancer progression. MiR-186-5p has been uncovered to be a suppressor in diversified cancers, including lung cancers. miR-186-5p suppresses cisplatin resistance by targeting SIX1 in NSCLC [Citation16]. miR-186 targets SIRT6 to retard lung cancer progression [Citation17]. In addition, lncRNA XIST sponges miR-186-5p to modulate NSCLC cell proliferation and invasion [Citation18]. Circ_0076305 combines miR-186-5p to strengthen cisplatin (DDP) resistance in NSCLC [Citation19]. Reports have focused on the interactions between circRNAs and miRNAs in lung cancer. However, the relationship between circCTNNB1 and miR-186-5p in lung cancer remains unknown.
This study was focused on studying the functions and regulatory mechanisms of competing endogenous RNAs (ceRNAs) of circCTNNB1 in lung cancer. This study uncovered that circCTNNB1 exhibited higher expression, resulted into poor prognosis in lung cancer, and formed the positive loop YY1/circCTNNB1/miR-186-5p/YY1 to aggravated lung cancer progression through the Wnt pathway. This study may provide new light for bio-markers in lung cancer treatment.
Materials and methods
Tissue samples
The lung cancer (n = 40) and adjacent normal (n = 40) tissues were collected from lung cancer patients at The Affiliated Tumor Hospital of Xinjiang Medical University, Xinjiang, China. The collected tissues were kept in liquid nitrogen right away for experiments. This study was acquired with the approval of the Ethics Committee of The Affiliated Tumor Hospital of Xinjiang Medical University (K-2022041). All lung cancer patients with no treatment have signed the informed consent.
Cell lines and culture
The normal bronchial epithelial cell line (BEAS-2B) and lung cancer cell lines (H1299, A549, PC9, and H1975) were bought from the American Tissue Culture Collection (ATCC, USA). The incubation of these cells in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, New York, NY, USA), including 10% foetal bovine serum (FBS, Gibco, USA), was performed in a wet incubator (5% CO2, 37°C).
RNase R and actinomycin D treatment
After RNase R (Sigma, Kawasaki, Kanagawa, Japan) treatment, the circular or linear CTNNB1 levels were examined by real-time quantitative polymerase chain reaction (RT-qPCR).
After actinomycin D (5 µg/mL, Sigma, Kawasaki, Kanagawa, Japan) treatment, at 0, 10, 20, and 30 h, the levels of circular or linear CTNNB1 were measured by RT-qPCR.
RT-qPCR
The RNAs from lung cancer tissues or cells were separated through TRIzol reagent (Invitrogen, USA). The transcription of RNAs to complementary DNA (cDNA) was made through the PrimeScript™ RT Master Mix kit (Takara, Dalian, China). Then, qRT-PCR was conducted through the SYBR Premix Ex Taq™ (Takara, Shanghai, China). The relative expressions were figured up through the 2−∆∆Ct method (GAPDH serves as the internal reference).
The primers were shown:
circ-CTNNB1:
Forward5’-TTTTCTTTTACATGCCCCCTCT-3,’
Reverse5’-GGAAGAAGAGAGTTTTTGTGTCCTT-3;’
miR-186-5p:
Forward5’-AAGAATTCTCCTTTTGGGCT-3,’
Reverse5’-GTGCGTGTCGTGGAGTCG-3;’
YY1:
Forward5’-GGAGGAATACCTGGCATTGACC-3,’
Reverse5’-CCCTGAACATCTTTGTGCAGCC-3;’
U6:
Forward5’-GCUUCGGCAGCACAUAUACUAAAAU-3,’
Reverse5’-CGCUUCACGAAUUUGCGUGUCAU-3;’
GAPDH:
Forward5’-GCCAAGGTCATCCATGACAAC-3,’
Reverse5’-ACCACTGACACGTTGGCAGTG-3;’
Subcellular fraction
H1299 and A549 cells were mixed with fractionation buffer and next centrifugated (500×g, 5 min). The supernatant (cytoplasm) and the precipitation (nucleus) were separated. CircCTNNB1 expression in the nucleus or cytoplasm was evaluated through RT-qPCR. GAPDH or U6 (for cytoplasm or nucleus) acted as an internal reference.
Cell transfection
The pcDNA3.1 targeting YY1 (pcDNA3.1/YY1) with negative control (Vector), shRNAs targeting circCTNNB1 (sh-circCTNNB1#1, #2, #3) or YY1 (sh-YY1) with negative control (sh-NC), miR-186-5p mimic with negative control (NC mimic) were constructed and purchased by GenePharma (Shanghai, China). The transfection of these plasmids into H1299 and A549 cells was made with Lipofectamine 2000 (Invitrogen, Waltham, MA, USA).
Cell counting kit-8 (CCK-8) assay
H1299 and A549 cells (1 × 104 cells/well) were plated into the 96-well plate. Each well was mixed with CCK-8 solution (10 μL, Dojindo Laboratories, Kumamoto, Japan) at 0, 24, 48, 72 and 96 h. Post incubation for another 4 h, one spectrophotometer (Thermo Fisher Scientific, MA, USA) was employed to measure the absorbance (450 nm).
Transwell assay
Transwell chambers pre-coated with (or without) Matrigel (BD Biosciences, MD, USA) were employed. The addition of serum-free DMEM medium was made into the upper chamber, and the addition of DMEM medium with 20% FBS was made into the lower chamber. The staining with crystal violet was conducted. The migrated and invaded cells were manually calculated through one microscope (Olympus Corporation, Tokyo, Japan).
SA-β-Gal assay
At pH 6.0, senescent cells had senescence-associated β-galactosidase (SA-β-Gal) activity. The artificial substrate X-Gal was utilized to stain cells to examine SA-β-Gal activity. The SA-β-Gal Kit (Beyotime Institute of Biotechnology, Shanghai, China) was employed. The cytoplasm (blue) stained with SA-β-Gal was deemed as positive.
Luciferase reporter assay
The pmirGLO dual-luciferase vectors (Promega, Madison, WI, USA) were inserted with wild-type or mutant-type sequences of circCTNNB1 (or YY1) to build the circCTNNB1 (or YY1)-WT and circCTNNB1 (or YY1)-MUT reporter vectors. H1299 and A549 cells were co-transfected with these above reporter vectors and miR-186-5p mimic/NC mimic through Lipofectamine 2000. After 48 h, the luciferase activity was evaluated through the dual-luciferase reporter assay system (Promega, Madison, WI, USA).
For evaluating the interaction ability between YY1 and the circCTNNB1 promoter, the pGL3 luciferase reporter vectors (Promega, Madison, WI, USA) were inserted with the circCTNNB1 promoter region (WT or MUT) to construct the circCTNNB1 promoter-WT and circCTNNB1 promoter-MUT reporter vectors. These constructed reporter vectors and pcDNA3.1/YY1 (or Vector) plasmids were co-transfected into H1299 and A549 cells.
RIP assay
H1299 and A549 cell lysates were mixed with magnetic beads conjugated with anti-Ago2, anti-IgG, or anti-YY1 antibodies. The negative control was the IgG group, and the positive control was the input group. After immunoprecipitation, the relative enrichment of circCTNNB1, miR-186-5p, and YY1 was assessed by RT-qPCR.
Western blot
The proteins were extracted from H1299 and A549 and were performed to 10% Sodium Dodecyl Sulphate-Polyacrylamide Gel Electrophoresis (SDS-PAGE). The moving of proteins to PVDF membranes (Beyotime, Shanghai, China) was done. After blocking, the PVDF membranes were added with the primary antibodies for 12 h at 4°C and the appropriate secondary antibodies for another 2 h. β-actin was set as the internal reference. The chemiluminescence detection kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA) was utilized to detect blots.
The primary antibodies were as follows: WNT3A (ab116222, 1 µg/mL, Abcam, China, Shanghai); β-catenin (ab223075, 1 µg/mL); c-myc (ab32072, 1/1000); YY1 (ab109228, 1/1000); β-actin (ab8226, 1 µg/mL).
In vivo assay
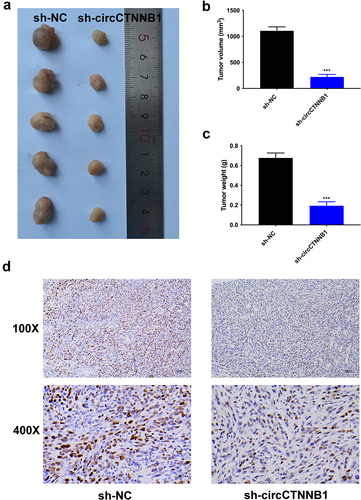
This assay was acquired with the approval of the Animal Care and Use Committee of Beijing Viewsolid Biotechnology Co. LTD (VS212601463). The male BALB/c nude mice (one-month-old, total n = 10, randomly n = 5 for each group) were obtained from the Vital River company (Beijing, China). The injection of transfected A549 cells (sh-NC, sh-circCTNNB1) into mice was performed. After 28 days, the mice were euthanized. The tumour size, volume, and weight were measured.
Immunohistochemistry (IHC) assay
The dewaxing and re-hydration for the 4 μm paraffin-embedded tumour tissue sections were done. After blocking, sections were cultured with Ki-67 antibodies at 4°C overnight and mixed with secondary antibodies (1:1000, ab7090, Abcam, Shanghai, China). The dyeing by diaminobenzidine (DAB) and re-dyeing by haematoxylin for sections were performed. Images were captured through one microscope (Nikon, Tokyo, Japan).
Statistical analysis
The data were displayed as the mean ± standard deviation (SD). The GraphPad Prism software, version 8.0 (GraphPad Software, La Jolla, CA), was utilized for statistical analysis. Each experiment was done in triplicate. The Student’s t-test (for two groups) or one-way analysis of variance (ANOVA, for multiple groups) was adopted to make comparisons. p < 0.05 was considered statistically significant.
Results
The higher expression of circCTNNB1 was discovered in lung cancer
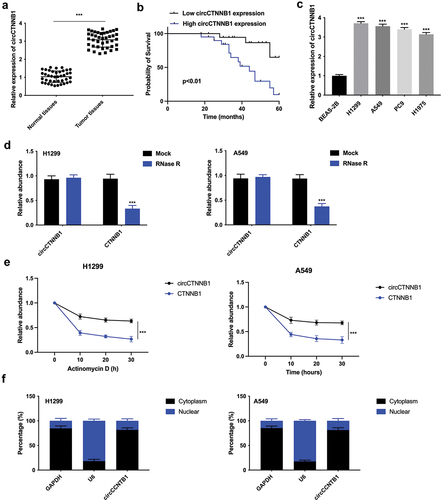
As shown in , circCTNNB1 expression was up-regulated in lung cancer tissues compared with the normal tissues. Higher circCTNNB1 expression resulted in poor prognosis in lung cancer patients (). circCTNNB1 exhibited higher expression in lung cancer cell lines (H1299, A549, PC9 and H1975) than in the bronchial epithelial cell line (BEAS-2B) (). Due to the higher expression of circCTNNB1 in H1299 and A549 cells, these two cell lines were collected for further experiments. The RNase R assay illustrated that CTNNB1 was degraded, whereas circCTNNB1 could resist RNase R digestion (). The half-life of circCTNNB1 was longer than that of CTNNB1 transcript after Actinomycin D treatment, indicating that circCTNNB1 was more stable than CTNNB1 (). These data indicated that circCTNNB1 was a circle RNA. Furthermore, circCTNNB1 was found to be located in the cytoplasm (). CircCTNNB1 expression was up-regulated in lung cancer.
Figure 1. The higher expression of circCTNNB1 was discovered in lung cancer.
CircCTNNB1 facilitated lung cancer cell proliferation, migration, invasion, and suppressed cell senescence
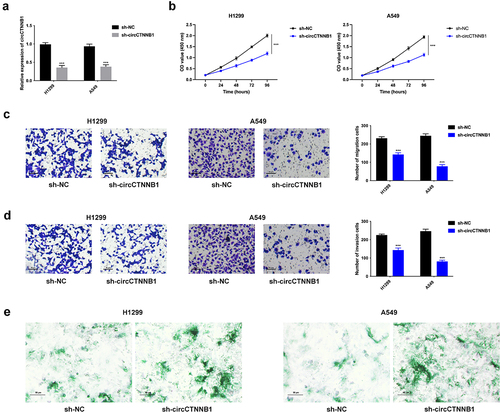
The knockdown efficiency of circCTNNB1 was confirmed in , and sh-circCTNNB1#1 was chosen for the next experiments due to its’ higher knockdown efficiency (sh-circCTNNB1). The cell proliferation was weakened after silencing circCTNNB1 (). The cell migration and invasion abilities were attenuated after circCTNNB1 knockdown (). The cell senescence was enhanced after circCTNNB1 suppression (). Taken together, circCTNNB1 facilitated lung cancer cell proliferation, migration, invasion, and suppressed cell senescence.
Figure 2. CircCTNNB1 facilitated lung cancer cell proliferation, migration, invasion, and suppressed cell senescence.
Knockdown of circCTNNB1 retarded the Wnt pathway
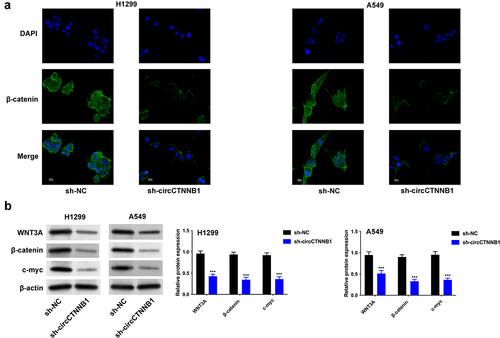
β-catenin transcribed by CTNNB1 is a key regulator in the Wnt pathway. So, circCTNNB1, which affects the Wnt pathway in lung cancer, needs further investigation. The nuclear translocation of β-catenin was decreased after circCTNNB1 inhibition (). The Wnt pathway-related proteins (WNT3A, β-catenin, and c-myc) were reduced after silencing circCTNNB1 (). The knockdown of circCTNNB1 retarded the Wnt pathway.
Figure 3. Knockdown of circCTNNB1 retarded the Wnt pathway.
CircCTNNB1 combined with miR-186-5p to target YY1
The related ceRNA mechanisms of circCTNNB1 were investigated. Under the screening condition (CLIP-seq Type: other CLIP), 13 miRNAs (miR-1294; miR-25-3p; miR-32-5p; miR-367-3p; miR-92b-3p; miR-363-3p; miR-223-3p; miR-944; miR-186-5p; miR-552-3p; miR-374b-3p; miR-668-3p; miR-526b-5p) were obtained. Through RT-qPCR, among these miRNAs, miR-186-5p was markedly elevated after circCTNNB1 knockdown (Figure S1). Thus, miR-186-5p was chosen for further experiments. Through the starBase online database, the binding sites between circCTNNB1 and miR-186-5p (YY1) were presented (). circCTNNB1 combined with miR-186-5p and miR-186-5p bound with YY1 through luciferase reporter and RIP assay (). The miR-186-5p expression was enhanced after circCTNNB1 knockdown, but YY1 mRNA and protein expression levels were reduced after circCTNNB1 knockdown or miR-186-5p mimic (). miR-186-5p expression was lower, and YY1 expression was higher in lung cancer tissues and cell lines (). circCTNNB1 combined with miR-186-5p to target YY1.
Figure 4. CircCTNNB1 combined with miR-186-5p to target YY1.
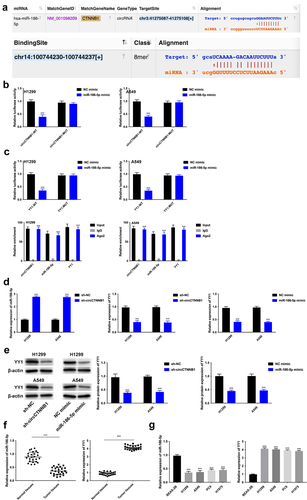
YY1 overexpression rescued the decreased cell proliferation, migration, invasion, and increased cell senescence mediated by circCTNNB1 suppression
The cell proliferation was weakened after circCTNNB1 knockdown, but this effect was reversed after YY1 overexpression (). In addition, the decreased cell migration and invasion stimulated by circCTNNB1 suppression were effectively alleviated after YY1 up-regulation (). The increased cell senescence induced by circCTNNB1 inhibition was relieved after YY1 overexpression (). These findings uncovered that YY1 overexpression could rescue the decreased cell proliferation, migration, and invasion and increased cell senescence mediated by circCTNNB1 suppression.
Figure 5. YY1 overexpression can rescue the decreased cell proliferation, migration, invasion, and the increased cell senescence mediated by circCTNNB1 suppression.
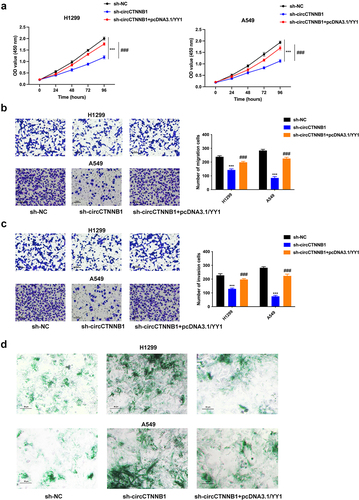
YY1 overexpression rescued the retarded Wnt pathway mediated by circCTNNB1 knockdown
The decreased nuclear translocation of β-catenin mediated by circCTNNB1 knockdown was rescued after YY1 amplification (). The reduced protein expressions of WNT3A, β-catenin, and c-myc triggered by circCTNNB1 inhibition were reversed after YY1 overexpression (). YY1 overexpression could rescue the retarded Wnt pathway mediated by circCTNNB1 knockdown.
Figure 6. YY1 overexpression can rescue the retarded Wnt pathway mediated by circCTNNB1 knockdown.
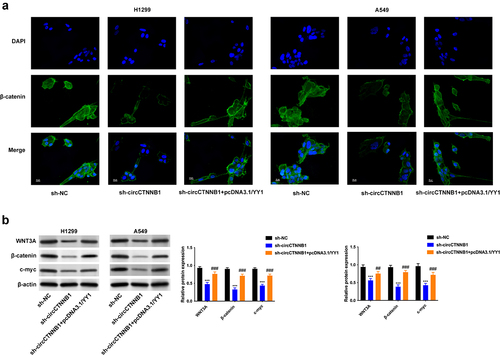
YY1 ranscriptionally activated circCTNNB1 to form YY1/circCTNNB1/miR-186-5p/YY1 loop
Whether YY1 can regulate the transcription of circCTNNB1 in lung cancer needs further investigation. The circCTNNB1 expression was positively correlated to YY1 expression in lung cancer tissues (). The expression of circCTNNB1 was reduced after YY1 inhibition (). The binding sites of YY1 are shown in . YY1 was verified to combine with the promoter of circCTNNB1 ().
Figure 7. YY1 can transcriptionally activate circCTNNB1 to form YY1/circCTNNB1/miR-186-5p/YY1 loop.
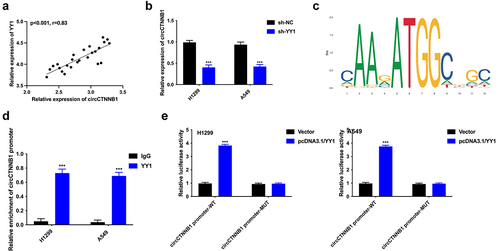
CircCTNNB1 accelerated tumor growth in vivo
The xenograft mouse models were adopted to determine the regulatory effects of circCTNNB1 on tumorigenesis in vivo. The A549 cells transfected with sh-circCTNNB1 (or sh-NC) plasmids were inoculated into mice. The tumour size, volume, and weight were reduced after circCTNNB1 suppression (). The Ki67 expression level was decreased after circCTNNB1 inhibition (). circCTNNB1 accelerated tumour growth in vivo.
Discussion
circRNAs exert regulatory functions in diversified ways, such as affecting proteins or miRNAs [Citation20,Citation21]. CircCTNNB1 has been discovered to participate in some diseases [Citation12–15], but its regulatory roles in lung cancer remain vague. In this study, the higher expression of circCTNNB1 was in lung cancer tissues and cell lines and resulted in poor prognosis. In addition, circCTNNB1 facilitated lung cancer cell proliferation, migration, invasion, and suppressed cell senescence.
The Wnt pathway has been disclosed to be a vital pathway in cancer progression [Citation22,Citation23]. The Wnt pathway exists in the regulation of lung cancer. lncRNA PKMYT1AR activates the Wnt pathway in non-small cell lung cancer to facilitate cancer stem cell maintenance [Citation24]. CircEIF3I affects the Wnt/β-catenin pathway to strengthen cell proliferation and invasion by targeting the miR-1253/NOVA2 axis in lung cancer [Citation25]. Cigarette smoking strengthens the Wnt/β-catenin pathway to stimulate tumorigenesis in lung cancer [Citation26]. Besides, MARVELD3 retards the Wnt/β-catenin pathway to suppress the epithelial-mesenchymal transition progress in non-small cell lung cancer [Citation27]. circCTNNB1 has been reported to activate Wnt signalling and up-regulate β-catenin protein expression [Citation14]. Besides, β-catenin transcribed by CTNNB1 is a key regulator in the Wnt pathway. So, circCTNNB1 affects the Wnt pathway in lung cancer and needs further investigation. Our results also revealed that the knockdown of circCTNNB1 retarded the Wnt pathway.
circRNAs affect cellular processes in cancers by sponging specific miRNAs to release mRNAs, forming a ceRNA axis [Citation28,Citation29]. For example, circ_0007401 sponges miR-6509-3p to target fli1, affecting gemcitabine resistance in pancreatic cancer [Citation30]. Furthermore, circ_0072088 regulates miR-1225-5p/WT1 axis to aggravate malignant behaviour in non-small cell lung cancer [Citation31]. Circ_0003747 targets the miR-338-3p/PLCD3 axis to intensify thyroid cancer progression [Citation32]. circSCN8A combines miR-1290 to release ACSL4, inhibiting malignant progression in non-small cell lung cancer [Citation33]. This ceRNA axis has also been confirmed to participate in lung cancer progression. However, the related ceRNA mechanisms of circCTNNB1 in lung cancer need more exploration. In this study, further mechanism-related experiments revealed that circCTNNB1 combined with miR-186-5p to target YY1. Through rescue assays, YY1 overexpression can rescue the decreased cell proliferation, migration, invasion, increased cell senescence, and the retarded Wnt pathway mediated by circCTNNB1 suppression.
YY1 is a common transcription factor that affects circRNA, lncRNA, or mRNA expression in cancers. YY1-induced circFIRRE contributes to the progression of osteosarcoma [Citation34]. YY1-stimulated up-regulation of lncRNA LINC00466 aggravates glioma progression through targeting miR-508/CHEK1 axis [Citation35]. Moreover, YY1-triggered lncRNA MCM3AP-AS1 targets the miR-340-5p/KPNA4 axis to strengthen angiogenesis in lung cancer [Citation36]. Besides, in tongue squamous cell carcinoma, YY1-mediated PTEN dephosphorylation relieves IR-stimulated DNA repair [Citation37]. Whether YY1 can regulate the transcription of circCTNNB1 in lung cancer needs further investigation. Similar to previous studies, YY1 can transcriptionally activate circCTNNB1, forming the YY1/circCTNNB1/miR-186-5p/YY1 loop. Besides, circCTNNB1 accelerated tumour growth in vivo.
A positive loop YY1/circCTNNB1/miR-186-5p/YY1 aggravates lung cancer progression through the Wnt pathway. This discovery may supply useful therapeutic bio-targets for lung cancer therapeutic strategies important in clinical diagnosis and treatment. This work also has some limitations for the influences of circCTNNB1 on lung cancer progression, such as lacking more human samples and more cellular processes (autophagy, exosomes, mitochondrial dysfunction, immune escape). In the future, more experiments will be carried out on lung cancer progression for further investigations.
Abbreviations
| competing endogenous RNAs | = | ceRNAs |
| non-small cell lung cancer | = | NSCLC |
| Circular RNAs | = | circRNAs |
| scavenger receptor class B type 1 | = | SRB1 |
Author contributions
YLS, YY, YYZ and CLL designed the study. YZ and SN collected the data. YYZ and CLL analysed the data. YLS and YY wrote the manuscript. All authors read and approved the final manuscript.
Supplemental Material
Download TIFF Image (56 MB)Cell STR file.zip
Download Zip (2.3 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available from the corresponding author.
Supplemental material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592294.2024.2369006
Additional information
Funding
References
- Collins LG, Haines C, Perkel R, et al. Lung cancer: diagnosis and management. Am Fam Physician. 2007 Jan 1;75(1):56–14.
- Nasim F, Sabath BF, Eapen GA. Lung cancer. Med Clin North Am. 2019 May;103(3):463–473. doi: 10.1016/j.mcna.2018.12.006
- Relli V, Trerotola M, Guerra E, et al. Abandoning the notion of non-small cell lung cancer. Trends Mol Med. 2019 Jul;25(7):585–594. doi: 10.1016/j.molmed.2019.04.012
- Lemjabbar-Alaoui H, Hassan OU, Yang YW, et al. Lung cancer: biology and treatment options. Biochim Biophys Acta. 2015 Dec;1856(2):189–210. doi: 10.1016/j.bbcan.2015.08.002
- Grivaux M, Zureik M, Marsal L, et al. Five-year survival for lung cancer patients managed in general hospitals. Rev Mal Respir. 2011 Sep;28(7):e31–8. doi: 10.1016/j.rmr.2008.07.001
- Li J, Sun D, Pu W, et al. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020 Apr;6(4):319–336. doi: 10.1016/j.trecan.2020.01.012
- Li F, Yang Q, He AT, et al. Circular RNAs in cancer: limitations in functional studies and diagnostic potential. Semin Cancer Biol. 2021 Oct;75:49–61. doi: 10.1016/j.semcancer.2020.10.002
- Fan Z, Bai Y, Zhang Q, et al. CircRNA circ_POLA2 promotes lung cancer cell stemness via regulating the miR-326/GNB1 axis. Environ Toxicol. 2020 Oct;35(10):1146–1156. doi: 10.1002/tox.22980
- Han J, Zhao G, Ma X, et al. CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem Biophys Res Commun. 2018 Sep 18;503(4):2429–2435. doi: 10.1016/j.bbrc.2018.06.172
- Zhang T, Wu DM, Luo PW, et al. CircNEIL3 mediates pyroptosis to influence lung adenocarcinoma radiotherapy by upregulating PIF1 through miR-1184 inhibition. Cell Death Dis. 2022 Feb 21;13(2):167. doi: 10.1038/s41419-022-04561-x
- Ishola AA, Chien CS, Yang YP, et al. Oncogenic circRNA C190 promotes non-small cell lung cancer via modulation of the EGFR/ERK pathway. Cancer Res. 2022 Jan 1;82(1):75–89. doi: 10.1158/0008-5472.CAN-21-1473
- Yang F, Liu Y, Xiao J, et al. Circ-CTNNB1 drives aerobic glycolysis and osteosarcoma progression via m6A modification through interacting with RBM15. Cell Prolif. 2023 Jan;56(1):e13344. doi: 10.1111/cpr.13344
- Chen C, Chang X, Zhang S, et al. CircRNA CTNNB1 (circCTNNB1) ameliorates cerebral ischemia/reperfusion injury by sponging miR-96-5p to up-regulate scavenger receptor class B type 1 (SRB1) expression. Bioengineered. 2022 Apr;13(4):10258–10273. doi: 10.1080/21655979.2022.2061304
- Qi J, Wang T, Zhang Z, et al. Circ-Ctnnb1 regulates neuronal injury in spinal cord injury through the Wnt/β-catenin signaling pathway. Dev Neurosci. 2022;44(3):131–141. doi: 10.1159/000521172
- Yang F, Fang E, Mei H, et al. Cis-Acting circ-CTNNB1 promotes β-Catenin signaling and cancer progression via DDX3-Mediated transactivation of YY1. Cancer Res. 2019 Feb 1;79(3):557–571. doi: 10.1158/0008-5472.CAN-18-1559
- Liu X, Zhou X, Chen Y, et al. miR-186-5p targeting SIX1 inhibits cisplatin resistance in non-small-cell lung cancer cells (NSCLCs). Neoplasma. 2020 Jan;67(1):147–157. doi: 10.4149/neo_2019_190511N420
- Ruan L, Chen J, Ruan L, et al. MicroRNA-186 suppresses lung cancer progression by targeting SIRT6. Cancer Biomark. 2018 Feb 6;21(2):415–423. doi: 10.3233/CBM-170650
- Wang H, Shen Q, Zhang X, et al. The long non-coding RNA XIST controls non-small cell lung cancer proliferation and invasion by modulating miR-186-5p. Cell Physiol Biochem. 2017;41(6):2221–2229. doi: 10.1159/000475637
- Wang X, Wang H, Jiang H, et al. Circular RNAcirc_0076305 Promotes Cisplatin (DDP) resistance of non-small cell lung cancer cells by regulating ABCC1 through miR-186-5p. Cancer Biother Radiopharm. 2023 Jun;38(5):293–304. doi: 10.1089/cbr.2020.4153
- Zhou WY, Cai ZR, Liu J, et al. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020 Dec 14;19(1):172. doi: 10.1186/s12943-020-01286-3
- Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79.
- Zhang Y, Wang X. Targeting the Wnt/β-catenin signaling pathway in cancer. J Hematol Oncol. 2020 Dec 4;13(1):165. doi: 10.1186/s13045-020-00990-3
- Duchartre Y, Kim YM, Kahn M. The Wnt signaling pathway in cancer. Crit Rev Oncol Hematol. 2016 Mar;99:141–149. doi: 10.1016/j.critrevonc.2015.12.005
- He Y, Jiang X, Duan L, et al. LncRNA PKMYT1AR promotes cancer stem cell maintenance in non-small cell lung cancer via activating Wnt signaling pathway. Mol Cancer. 2021 Dec 2;20(1):156. doi: 10.1186/s12943-021-01469-6
- Chen T, Feng G, Xing Z, et al. Circ-EIF3I facilitates proliferation, migration, and invasion of lung cancer via regulating the activity of Wnt/β-catenin pathway through the miR-1253/NOVA2 axis. Thorac Cancer. 2022 Nov;13(22):3133–3144. doi: 10.1111/1759-7714.14665
- Malyla V, Paudel KR, De Rubis G, et al. Cigarette smoking induces lung cancer tumorigenesis via upregulation of the WNT/β-catenin signaling pathway. Life Sci. 2023 Aug 1;326:121787. doi: 10.1016/j.lfs.2023.121787
- Li S, Qi S, Li Y, et al. MARVELD3 inhibits the epithelial-mesenchymal transition and cell migration by suppressing the Wnt/β-catenin signaling pathway in non-small cell lung cancer cells. Thorac Cancer. 2023 Apr;14(12):1045–1058. doi: 10.1111/1759-7714.14844
- Liang ZZ, Guo C, Zou MM, et al. circRNA-miRNA-mRNA regulatory network in human lung cancer: an update. Cancer Cell Int. 2020;20(1):173. doi: 10.1186/s12935-020-01245-4
- Tang X, Ren H, Guo M, et al. Review on circular RNAs and new insights into their roles in cancer. Comput Struct Biotechnol J. 2021;19:910–928. doi: 10.1016/j.csbj.2021.01.018
- Han C, Zheng H, Hu D, et al. Hsa_circ_0007401 regulates gemcitabine resistance of pancreatic cancer through the hsa-miR-6509-3p/fli1 axis. Medicine (Baltimore). 2023 May 26;102(21):e33775. doi: 10.1097/MD.0000000000033775
- Zhu X, Wan J, You X, et al. Circular non-coding RNA circ_0072088 serves as a ceRNA, targeting the miR-1225-5p/WT1 axis to regulate non-small cell lung cancer cell malignant behavior. Thorac Cancer. 2023 May 23;14(20):1969–1979. doi: 10.1111/1759-7714.14943
- Dou XL, Xia FD, Li XY. Circ_0003747 promotes thyroid cancer progression by sponging miR-338-3p to upregulate PLCD3 expression. Epigenetics. 2023 Dec;18(1):2210339. doi: 10.1080/15592294.2023.2210339
- Liu B, Ma H, Liu X, et al. CircSCN8A suppresses malignant progression and induces ferroptosis in non-small cell lung cancer by regulating miR-1290/ACSL4 axis. Cell Cycle. 2023 Apr;22(7):758–776. doi: 10.1080/15384101.2022.2154543
- Yu L, Zhu H, Wang Z, et al. Circular RNA circFIRRE drives osteosarcoma progression and metastasis through tumorigenic-angiogenic coupling. Mol Cancer. 2022 Aug 19;21(1):167. doi: 10.1186/s12943-022-01624-7
- Li F, Shen ZZ, Xiao CM, et al. YY1-mediated up-regulation of lncRNA LINC00466 facilitates glioma progression via miR-508/CHEK1. J Gene Med. 2021 Jan;23(1):e3287. doi: 10.1002/jgm.3287
- Li X, Yu M, Yang C. YY1-mediated overexpression of long noncoding RNA MCM3AP-AS1 accelerates angiogenesis and progression in lung cancer by targeting miR-340-5p/KPNA4 axis. J Cell Biochem. 2020 Mar;121(3):2258–2267. doi: 10.1002/jcb.29448
- Zhao L, Li R, Qiu JZ, et al. YY1-mediated PTEN dephosphorylation antagonizes IR-induced DNA repair contributing to tongue squamous cell carcinoma radiosensitization. Mol Cell Probes. 2020 Oct;53:101577. doi: 10.1016/j.mcp.2020.101577