ABSTRACT
Gametogenetin binding protein 2 (GGNBP2) was indispensable in normal spermatids for transformation into mature spermatozoa in mice, and when Gametogenetin binding protein 2 is bound to BRCC36 and RAD51, the complex participates in repairing DNA double-strand breaks (DSB) during the meiotic progression of spermatocytes. Ggnbp2 knockout resulted in the up-regulation of H2AK119ubi and down-regulation of H2BK120ubi in GC-2 cells (mouse spermatogonia-derived cell line) and postnatal day 18 testis lysate. Our results also demonstrated that Gametogenetin binding protein 2 inducedASXL1 to activate the deubiquitinating enzyme BAP1 in deubiquitinating H2A, while Gametogenetin binding protein 2 knockout disrupted the interaction between ASXL1 and BAP1, resulting in BAP1 localization change. Furthermore, the Gametogenetin binding protein 2 deletion reduced H2B ubiquitination by affecting E2 enzymes and E3 ligase binding. Gametogenetin binding protein 2 regulated H2A and H2B ubiquitination levels and controlled H3K27 and H3K79 methylation by PRC2 subunits and histone H3K79 methyltransferase. Altogether, our results suggest that Ggnbp2 knockout increased DNA damage response by promoting H2A ubiquitination and H3K27trimethylation (H3K27me3) and reduced nucleosome stability by decreasing H2B ubiquitination and H3K79 dimethylation (H3K79me2), revealing new mechanisms of epigenetic phenomenon during spermatogenesis. Gametogenetin binding protein 2 seems critical in regulating histone modification and chromatin structure in spermatogenesis.
Introduction
GGNBP2, also termed dioxin-inducible factor-3 (DIF-3), was first reported as a target gene mediating reproductive toxicity from dioxin, an environmental toxic agent [Citation1]. It has been reported to be involved in many types of cancers, including breast cancer [Citation2], laryngeal carcinoma [Citation3], ovarian cancer [Citation4,Citation5], glioma [Citation6] and prostate cancer [Citation7]. GGNBP2 is highly expressed in the adult testis, and its expression is tightly associated with spermatogenesis [Citation8]. Our previous study revealed that GGNBP2 plays a crucial role in maintaining the structural integrity of the adluminal germ epithelium and the transformation of spermatids into mature spermatozoa in mice. Therefore, the loss of GGNBP2 function may result in defects in these processes and could potentially lead to male infertility [Citation9]. During spermiogenesis, extensive histone modifications have been reported to occur in developing haploid spermatids, as well as morphological alterations of the genetic material to form compact nuclei. These distinct observations attracted our attention to investigate the role of some histone-modifying enzymes or histone modification in the reaction.
Histone modifications such as acetylation, methylation, phosphorylation and ubiquitination are important for chromatin structure alteration and gene expression [Citation10,Citation11]. Histone ubiquitination is unique among histone modifications due to the relatively bulky size of ubiquitin and has previously been shown to stimulate or repress various cellular processes [Citation12]. Histone H2A and H2B are mainly monoubiquitylated and are important in regulating gene expression and nucleosome stabilization [Citation13]. Ubiquitination of the histone H2A Lys-119 residue, the first identified ubiquitin conjugate, is primarily catalysed by polycomb repressive complex 1 (PRC1) [Citation14] and has been linked to gene repression. The process of ubiquitination is conducted by the sequential action of activating (E1), conjugating (E2) and ligating (E3) enzymes and can be reversed by deubiquitinases (DUBs) [Citation15]. The balance between ubiquitination and deubiquitination regulates various biological processes, including spermatogenesis [Citation16]. The roles of DUBs in the modulation of DNA repair and DNA damage signalling are well established [Citation17]. Feng et al. reported that BRCC36 affected the number of Lys(63)-linked ubiquitin chains at chromatin, with H2A as one of its substrates [Citation18]. BRCC36 was also shown to promote BRCA1 function in DNA damage response and played an instrumental role in limiting DNA end resection and fine-tuning homologous recombination (HR) repair [Citation19]. Although we showed that GGNBP2 or GGN bonded to BRCC36 and RAD51 to induce DNA DSB repair in our previous study [Citation20], it remains unclear whether GGNBP2 can regulate H2A ubiquitination by forming a complex with other DUBs.
H2B lysine 120 monoubiquitylation (H2Bk120 or H2Bubi) is mediated by human RNF20/RNF40 and UbcH6 and is required for active transcription of genes, with its level significantly increasing during differentiation [Citation18,Citation21]. The ubiquitin-conjugating enzyme E2B (UBE2B; also known as Rad6b or HR6B) is one of the two mammalian homologs of the Saccharomyces cerevisiae E2 enzyme named RAD6/UBC2 [Citation22]. In yeast, Rad6 works with Bre1, an E3 ligase, to catalyse the monoubiquitylation of histone H2Bk123 on a C-terminal lysine residue [Citation23]. In humans, two homologs of yRAD6 (hRad6a and hRad6b) [Citation22] and two homologs of yBRE1 (RNF20 and RNF40) [Citation24] were shown to participate in histone H2Bk120 ubiquitylation. Additionally, it was reported that male mice with Hr6a knockout were fertile while those with Hr6b knockout were infertile. In humans, HR6B (UBE2B) mutations were also reported to be associated with male infertility [Citation25]. Further, RNF20-mediated H2B ubiquitination regulates meiotic recombination by promoting chromatin relaxation. In vivo studies indicated that H2Bub could stabilize nucleosomes [Citation26] and facilitate nucleosome reassembly during transcription elongation [Citation27]. Thus, H2Bubi has a complex role in modulating nucleosome and chromatin structure.
Gametogenetin binding protein 2 (GGNBP2), comprising 697 amino acids with a molecular weight of approximately 79 kD, is a highly conserved gene localized to the tumour-suppressor locus D17S800-D17S930andalso known to be involved in spermatogenesis [Citation27]. Presently, the epigenetic mechanisms that involve H2AK119ubi and H2BK120ubi in Ggnbp2 deletion-related azoospermia are poorly understood. In this present study, we investigated the role of GGNBP2 in regulating H2A and H2B ubiquitination and H3 methylation. Our findings suggest that GGNBP2 stabilized nucleosomes by targeting UBE2B and was involved in the E3 enzyme RNF40-mediated monoubiquitination of histone H2B. Moreover, we also demonstrated that GGBNP2 played a key role in regulating trans-histone crosstalk by modulating H2A and H2B ubiquitination and H3 methylation via the activity of Polycomb repressive complex 2 (PRC2) subunits and Dot1-like protein (DOT1L). Altogether, these findings shed new light on the molecular mechanisms underlying Ggnbp2 deletion-related azoospermia, suggesting that epigenetic dysregulation could be a potential contributor to male infertility.
Materials and methods
Mice and cell line
A total of 30 mice were tested, and 10 Ggnbp2 knockout mice were obtained. Briefly, Ggnbp2 knockout (Ggnbp2KO) heterozygous female mice with C57BL/6 background were crossed with wild-type (WT) male mice of 129/Sv background. Then, these Ggnbp2 heterozygous males and females with mixed backgrounds were bred. Mouse tail genomic DNA was extracted using Zymo Research Corp (Irvine, CA) genomic DNA-tissue mini prep kits following the manufacturer’s instructions (Zymo Research Corp, Irvine, CA) and genotyped by polymerase-chain reaction (PCR) using primer sets as previously reported [Citation28]. The animals were housed under 12-h day and night cycles, with free access to food and water. The Sertoli cell-only testis model was generated by a single intraperitoneal busulfan injection of 30 mg/kg into adult WT and mutant mice [Citation29]. The animal studies were approved by the Animal Care and Use Committee of the First Hospital of Jilin University. The mice were sacrificed under ketamine anaesthesia, and all efforts were made to minimize their discomfort.
Mouse spermatocyte-derived GC-2 cells (GC-2 cells) were purchased from the American Tissue Culture Collection (ATCC, Manassas, VA, USA). They were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM, Sigma, St Louis, MO, USA) supplemented with 10% heat-inactivated foetal bovine serum (FBS; Sigma) and 1% penicillin-streptomycin (Invitrogen, San Francisco, CA, USA) at 37°C in a humidified incubator containing 5% CO2 and 95% air atmosphere.
Generation of stable Ggnbp2KO GC-2 cells
Ggnbp2 double nickase plasmids (sc -432,161-NIC) were purchased from Santa Cruz Biotech (Santa Cruz, CA, USA). One day before Ggnbp2 plasmid transfection, GC-2 cells were seeded into a 6-well plate at 2 × 105 cells per well. When the confluency reached 70%, the growth medium was replaced with fresh antibiotic-free growth media (Santa Cruz Biotech). Then, 300 μL plasmid DNA/UltraCruz transfection reagent complexes (Santa Cruz Biotech) were added to each well, followed by overnight incubation. The medium was then replaced with the growth medium 24 h after transfection. Stably transfected cells were obtained after treatment with 4 μg/mL puromycin (Sigma) for 2 weeks. Then, GFP-positive and puromycin-resistant clones were digested using trypsin (Sigma) and transferred into 96-well plates, with 2–3 cells dilution per well. The procedure of single-cell cloning was repeated three times to obtain 100% clonal purity. Reverse-transcription PCR (RT-PCR) and Western blotting were then used to verify the effectiveness of the stable Ggnbp2KO cell lines.
Transient transfection of Ggnbp2 overexpression plasmids or siRNA
The human Ggnbp2 complementary DNA (cDNA) fragment was obtained by PCR using human testis cDNA as the template and validated by DNA sequencing. The correct full-length human Ggnbp2 cDNA was cloned into the pcDNA3-6HisC vector (Invitrogen) to construct the mammalian expression plasmid pcDNA3-HisC tagged-Ggnbp2 (pcDNA3–Ggnbp2). For transfecting Ggnbp2 overexpressing plasmid into GC-2 cells, the cells were seeded at a density of 2 × 105 per well in a 6-well plate and cultured in DMEM to approximately 70% confluence. Small interfering RNA (siRNA) transfection was performed using the siRNA Transfection Reagent (Santa Cruz Biotech, sc -29,528), following the manufacturer’s instructions. Plasmid and siRNA transfection was performed with the Trans IT-X2 dynamic delivery system (Mirus Bio, Madison, WI, US) as per the manufacturer’s instructions. ASXL1 (sc -145,389), UBE2B (sc -154,849), RNF40 (sc -153,050) and scramble (sc -37,007) siRNA were purchased from Santa Cruz Biotech (Santa Cruz, CA). The medium was replaced with DMEM containing 10% FBS, and the cells were cultured in the same medium for 72 h, then harvested for Western blotting.
Micrococcal nuclease (MNase) digestion of chromatin
The assay was performed as previously described with minor modifications [Citation26,Citation28]. Briefly, a germ cell suspension was made for the testes. The decapsulated testes were incubated in PBS containing 0.5 mg/mL collagenase (type IV) for 15 min at 32°C in a water bath placed on a shaker. After washing twice with PBS containing 1.0 µg/mL DNase, 1.0 µg/mL trypsin in PBS was added and incubated for 15 min at 32°C. The suspension was filtered through an 80 µm nylon mesh (50 µm for 30 days old). The filtrate was washed twice with PBS and incubated in a hypotonic buffer (10 mMHepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol and 1 mMdithiothreitol [DTT]) supplemented with 0.1% Triton X-100 for 30 min. When GC-2 cells’ growth reached 90% confluency, they were harvested, washed twice with PBS and incubated in a hypotonic buffer. Then, the suspension was subjected to 12 strokes in a Dounce homogenizer fitted with pestle B. The lysate was centrifuged at 3,000 rpm, and 4°C to remove cytomembrane debris, and the resulting supernatant was centrifuged at 13,000 rpm. The nuclei-containing pellet was resuspended in a reaction buffer (50 mMTris-HCL, 5 mM CaCl2, 100 µg/µL BSA, pH 7.9). The total amount of nuclei obtained was determined using an aliquot resuspended in 1 M NaOH, and the optical density was measured at 260 nm and normalized to a DNA concentration of ~1.5 mg/mL.
Equal amounts of nuclei were split into four equal aliquots. An aliquot of the nuclei was set aside as mock digestion (0 U/mL or 0 min). The MNase stock (Kunitz unit; 20 KU/µL) was serially diluted, added to the nuclei to a final enzyme concentration of 0.2, 0.4 and 0.6 KU/µL, and digested for 5 min at 37°C. Alternatively, the nuclei were treated with MNase (0.4 KU/µL) at indicated incubation time (1, 3, 4 and 5 min), to which an equal volume of 2×TNESK buffer (20 mMTris – HCl, pH 7.4, 0.2 M NaCl, 2 mM EDTA, and 2% SDS) and proteinase K (0.2 mg/mL) was added. The DNA was then purified twice using phenol-chloroform extraction, followed by ethanol precipitation. The DNA was separated by electrophoresis in a 1.2% agarose gel and stained with ethidium bromide for visualization. DNA recovery following MNase digestions was monitored using a spectrophotometer.
Differential salt solubility assay
GC-2 cell nuclei were isolated and measured by spectrophotometry, as described above. Equal amounts of nuclei were partitioned into five equal aliquots. The nuclei were washed by gentle resuspension using wide orifice tips in extraction buffer (20 mMTrisCl [pH 7.5], 0.25% Triton X-100, 1 Complete-EDTA free protease inhibitor mixture [Roche, Mannheim, Germany]) containing increasing concentrations of NaCl. After incubating on ice for 5 min, the nuclei were centrifuged at 13,000 rpm for 5 min to obtain soluble fractions. Then, they were resuspended in 1×Lammeli sample buffer and subjected to Western blotting using histone H3.
Nuclear and cytosolic fractionation
Testicular germ cell suspension was prepared on postnatal day 18 (P18). The Thermo Scientific NE-PER Nuclear and Cytoplasmic Extraction Kit has been used for efficient cell lysis and extraction of separate cytoplasmic and nuclear protein fractions, according to manufacturer’s instructions (Thermo Fisher Scientific Inc., Rockford, IL USA).
Chromosome spread and immunofluorescence staining
The testes of mutant and WT mice aged postnatal day 28 (P28) were removed and processed as previously described [Citation30]. Briefly, after removal, the testes were decapsulated into hypotonic sucrose extraction buffer (30 mMTris, 50 mM sucrose, 17 mMtrisodium citrate dihydrate, 5 mMethylenediaminetetraacetic acid [EDTA], 0.5 mMdithiothreitol (DTT), and 0.5 mMphenylmethylsulphonyl fluoride [PMSF], pH 8.2) for 30 min, and mixed in a ratio of 1:2 with 100 mM sucrose just before spreading on slides preincubated with 4% paraformaldehyde. The slides were incubated in a humid chamber for 1 h and dried for 30 min. Lastly, the slides were washed twice for 2 min in 0.4% Photo-Flo (Kodak, Rochester, US) and dried at room temperature or either stained or stored at −80°C.
For immunofluorescence, the spread preparations were rinsed 3 times for 5 min in PBS, blocked in 5% BSA for 1 h, and incubated overnight at 4°C with the corresponding primary antibodies. Incubations with secondary antibodies were performed at room temperature in the dark for 60 min. The secondary antibodies used were TRITC-conjugated donkey anti-mouse immunoglobulin G (IgG), FITC-conjugated donkey anti-rabbit IgG, TRITC-conjugated donkey anti-rabbit IgG and FITC-conjugated donkey anti-mouse IgG (1:100, Jackson ImmunoResearch Lab, West Grove, PA, USA). The nuclei were stained with DAPI (2 mg/mL) during the final washing step. Then, the images were immediately collected using an Olympus fluorescence microscope (IX71/DP72, Tokyo, Japan).
Western blotting
This experiment was performed as previously described [Citation28]. Briefly, the testes were decapsulated and homogenized for 30 s using a Tissue-Tearor (RPI Corp, Mt. Prospect, IL) in a RIPA buffer containing 50 mMTris-HCl, 150 mMNaCl, 1% NP-40, 1 mM EDTA, 2 mM MgCl2, 0.5% sodium deoxycholate, complete protease inhibitor and phosphorylase inhibitor cocktail (Roche, Mannheim, Germany). GC-2 cells were harvested and washed in PBS 3 times and prepared in RIPA buffer. Then, the lysate was sonicated, resolved by SDS/PAGE gels, and then electroblotted to PVDF membranes (Millipore). After blocking with 3% non-fat milk in Tris-Buffered Saline Tween-20 (TBST), the membranes were incubated with their corresponding primary antibody followed by HRP-conjugated secondary antibodies. Signals were detected using a Pierce ECL western blotting substrate detection kit (Pierce) (Table S1). All membranes were reblotted with β -Actin or GAPDH antibody as the loading control. The intensity of specific bands was scanned using the TotalLab Quant 12 software image analysis. The results are presented as the ratio of target protein over β-Actin or GAPDH.
Immunoprecipitation (IP)
After protein concentration was determined, equal amounts of testis extracts (200 µg) were added to 20 µL protein A/G Plus-agarose beads (Santa Cruz) for 1 h at 4°C with agitation on a rotator to remove the non-specific interacting proteins. Following centrifugation, the supernatant was then incubated with 1 μg anti-GGNBP2, 1 μg anti-ASXL1, 1 μg anti-UBE2B, or 1 μg anti-RNF40 for 4 h at 4°C on a rotator to allow co-IP. Then, 20 μL of Protein A/G PLUS-Agarose was added to each sample, and they were incubated overnight to precipitate the immunocomplexes. Normal rabbit IgG was used as the negative control. The next day, the samples were washed three times with buffer A (50 mMTris-HCl, 0.5% Triton X-100, 1 mM EDTA, 150 mMNaCl, pH = 8.0), and one time with buffer B (10 M Tris-HCl, 0.1% Triton X-100, pH = 7.5). Bound protein complexes were boiled with 2× sample buffer and separated on SDS-PAGE. The proteins were then electroblotted onto a PVDF membrane and immunoblotted with the corresponding antibodies.
Statistical analysis
All statistical analyses were performed with the Instat program version 3.06 (GraphPad Software, San Diego, CA, USA). All experiments were repeated at least three times. Statistical analyses were performed using the Statistical Package for the Social Sciences (SPSS) software (v17.0, IBM Inc., Chicago, IL, USA). All experiments were performed at least three times. The data are presented as means ± SEM. For comparisons among different groups, one-way analysis of variance (ANOVA) was used, and for comparisons between two different groups, the student’s t-test was used. A P-value < .05 was considered statistically significant.
Results
Ggnbp2 knockout affects GC-2 cell chromatin compaction and nucleosome stability
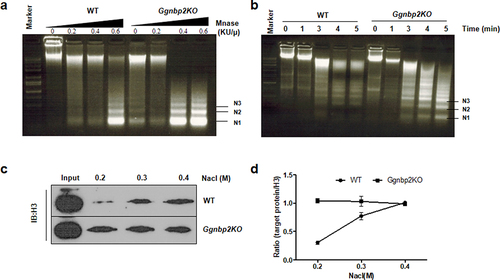
In this study, the immortalized GC-2 cell line, originally from mouse spermatocytes, was used to assess the effects of Ggnbp2 knockout on chromatin condensation and nucleosome stability. To assess whether such alterations occur as a result of Ggnbp2 loss, Stable Ggnbp2KO GC-2 cells were established using CRISPR/Cas9, and the degree of chromatin compaction in WT and Ggnbp2KO cells was examined with micrococcal nuclease (MNase) digestion, which cuts linker DNA that connects the nucleosomes and detects the chromatin compaction status (). The results showed that whole nuclei isolated from immortalized Ggnbp2KO cells were more thoroughly digested than those from WT at 0,0.2, 0.4 and 0.6 KU/µL MNase treatment () or within 1–5 min of treatment with 0.6 KU/µL MNase (). Next, we performed a histone solubility assay using increasing salt concentrations to examine the effects of Ggnbp2 knockout on nucleosome stability. We observed that high salt concentrations could increase histone solubility, and when the concentration reached 2.0 M, the relative soluble H3 amounts were similar between the two groups (Supplement Figure s1A), and they were used as control to show that equal amounts of nuclei were used in the assay (). Our results showed that H3 was more soluble at very low salt concentrations (0.2–0.4 M) in Ggnbp2KO cells compared with WT, which highly suggested that Ggnbp2 deletion impacted the stable binding of histones with chromatin.
Figure 1. Ggnbp2 controls both chromatin compaction and nucleosome stability.
Ggnbp2 deletion leads to the up-regulation of H2AK119ubi and down-regulation of H2BK120ubi
In spermiogenesis, extensive epigenetic modifications occur during differentiation for non-functional sperm to become mature spermatozoa [Citation31]. To assess whether Ggnbp2 affects nucleosome stability via epigenetic mechanisms, we evaluated the protein expression of histones H2AK119ubi and H2BK120ubi in vitro using GC-2 cells and in vivo using postnatal day-18 testes lysate. To enrich the spermatocyte population and avoid the presence of post-meiotic germ cells, postnatal day-18 testes were chosen for Western blotting. The in vitro results showed a differential expression pattern of histone H2AK119ubi and H2BK120ubi either in GC-2 cells or postnatal day 18 testis lysate (). In Ggnbp2KO cells, based on the reactivity scored with antibodies against histone H2A and histone H2B, we observed an increase in H2AK119ubi levels while H2BK120ubi levels decreased (). To exclude the possibility of sertoli cell contamination at histone ubiquitination level, we generated a sertoli cell-only model by a single intraperitoneal busulfan injection of 30 mg/kg into adult WT and mutant mice. Western blotting revealed no significant difference in H2AK119ubi and H2BK120ubi levels between the two groups, which suggested the differential expression pattern of histone H2AK119ubi and H2BK120ubi occurred in germ cells (Supplement Figure S1B-D), not in sertoli cells.
Figure 2. Ggnbp2 deletion leads to the up-regulation of H2AK119ubi and the down-regulation of H2BK120ubi.
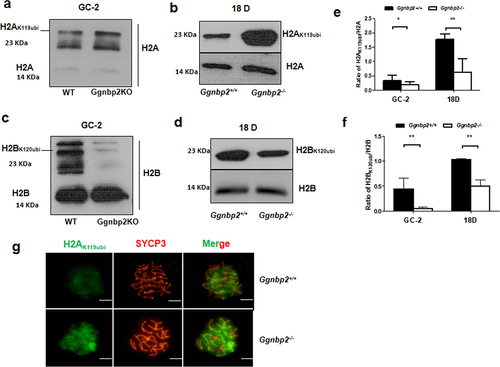
We also analysed the H2AK119ubi staining pattern in the spread nuclei of testes in WT and Ggnbp2KO 4-week-old male mice. Similar to the study of Baarends et al. [Citation32], we observed intense staining of ubiquitin in the XY body and lower overall staining of the nucleus due to the presence of other ubiquitinated proteins in mouse spermatocytes, while much more intense staining was observed in the XY body of Ggnbp2KO spermatocytes ().
GGNBP2 interacts and colocalizes with ASXL1 but not BAP1
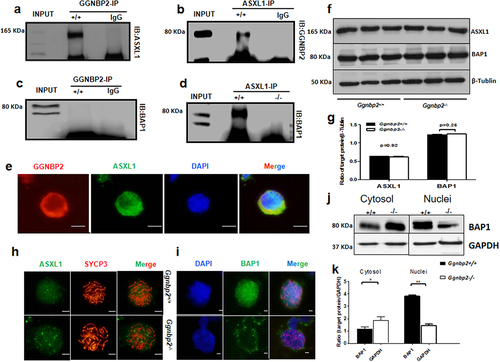
Since Ggnbp2 loss could increase H2AK119ubi expression, its underlying mechanism was investigated. UCH-class DUB BAP1 and its activator ASXL1 are the best characterized deubiquitinating enzyme specific for H2AK119ubi. BAP1/ASXL1 forms the PR-DUB complex, which was shown to deubiquitinateH2A [Citation33]. IP was performed to determine whether endogenous GGNBP2 could bind to these specific deubiquitinating enzymes to deubiquitinate H2A. Our results showed that GGNBP2 could be co-immunoprecipitated with ASXL1 but not with BAP1 (). In addition, ASXL1 was also co-immunoprecipitated with BAP1 (). Next, we examined GGNBP2 and ASXL1 expression in the spermatocytes of mice via immunofluorescence using a testicular cell suspension spun down onto microscope slides. We found that both GGNBP2 and ASXL1 expressed in the nucleus and cytoplasm of spermatocytes colocalized very well with each other (). These results suggest GGNBP2 acted as a scaffold protein to facilitate ASXL1 to activate the deubiquitinating enzyme BAP1 in deubiquitinating H2A.
Figure 3. GGNBP2 interacts and colocalizes with ASXL1. The absence of GGNBP2 affects ASXL1 and BAP1 interaction as well as BAP1 translocation.
GGNBP2 loss disrupts ASXL1 and BAP1 interaction resulting in BAP1 localization change
BAP1 requires activation by an N-terminal fragment of Polycomb group protein ASX, a protein necessary for the long-term repression of HOX genes, to deubiquitinateH2A [Citation34]. Here, we found that ASXL1 was not associated with BAP1 in the testis when the Ggnbp2 was knocked out, although Western blotting analysis revealed that neither ASXL1 nor BAP1 protein expression was affected in Ggnbp2KO total testis (). To demonstrate the association between GGNBP2 and ASXL1, immunofluorescence of spermatocyte chromosome spread was used to determine whether the location of ASXL1 or BAP1 was changed due to GGNBP2 loss. In WT cells, the levels of ASXL1 and BAP1 were found to be high in pachytene spermatocyte nuclei, the subcellular compartment where the ASXL1/BAP1 complex regulated H2A deubiquitination (). In Ggnbp2 knockout spermatocytes, we observed interesting alterations in the pattern of BAP1. No obvious BAP1 staining was detected in the nucleus compared with the cytoplasm. Western blotting analysis confirmed Ggnbp2 knockout disrupted BAP1 entry into the nucleus (). The results showed that GGNBP2 loss reduced deubiquitinase activity by disrupting ASXL1/BAP1 binding and preventing BAP1 from entering the nucleus.
Ggnbp2 knockout affects ubiquitin-conjugating E2 enzymes UBE2B and ubiquitin E3 ligase RNF40 interaction
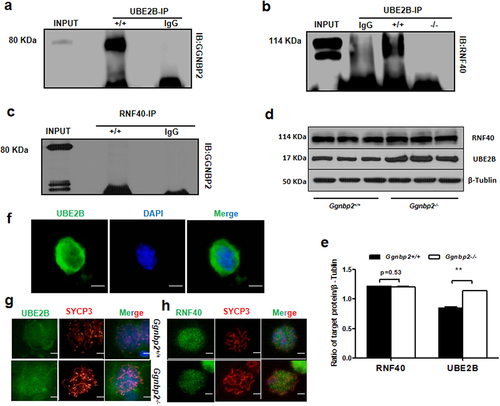
The ubiquitin-conjugating E2 enzymes UBE2A (HR6A) and UBE2B (HR6B) are two very similar mammalian homologs of yeast Rad6 [Citation22]. When bonded with the E3 enzyme BRE1 (RNF20 and/or RNF40), it regulates transcription by catalysing the monoubiquitination of histone H2B at Lys-120 to form H2BK120ubi. Our binding analyses revealed a specific interaction between GGNBP2 and UBE2B () and between UBE2B and RNF40 () but not between GGNBP2 and RNF40 (). In addition, GGNBP2 deletion was found to disrupt the binding of UBE2B with RNF40, accompanied by increased expression of UBE2B (). Next, we investigated the impact of GGNBP2 deficiency on the localization of UBE2B and RNF40 in pachytene spermatocytes. Immunofluorescence showed no difference in the staining patterns between the two groups (). Overall, these experiments revealed that deletion of GGNBP2 reduced H2B ubiquitination by affecting E2 enzymes and E3 ligase binding, recruiting less ubiquitin protein to H2B at Lys-120.
Figure 4. GGNBP2 interacts and colocalizes with UBE2B, and GGNBP2 loss affects UBE2B and RNF40 interaction.
GGNBP2 regulates H2A and H2B ubiquitination levels
Since GGNBP2 affected in vivo H2A and H2B ubiquitination by interacting with ASXL1 and UBE2B, respectively, we investigated the in vitro mechanism by which GGNBP2 regulates H2A and H2B ubiquitination using RNA interference (RNAi). First, we transfected Ggnbp2KO GC-2 cells with Ggnbp2 overexpressed plasmid. As shown in Ggnbp2 overexpression restored H2A and H2B ubiquitination levels. Then, we transfected WT and Ggnbp2KO cells with control scrambled siRNA and ASXL1 siRNA. The knockdown efficiency of siRNA was verified by Western blotting for ASXL1. As expected, a significant reduction in ASXL1 level was observed in both cell strains and increased H2AK119ubi expression (). To further confirm that GGNBP2 loss could affect the activation of BAP1 by ASXL1 in deubiquitinating H2A, Ggnbp2KO cells were transfected with ASXL1 siRNA together with Ggnbp2 overexpression plasmids. Western blotting with H2AK119ubi antibody indicated that overexpressing Ggnbp2 did not restore the binding between ASXL1 and BAP1, and deubiquitinase BAP1 was not activated (). These results highlight the potential roles of GGNBP2 in H2AK119 ubiquitination.
Figure 5. GGNBP2 regulates histone H2A and H2B ubiquitination levels in vitro.
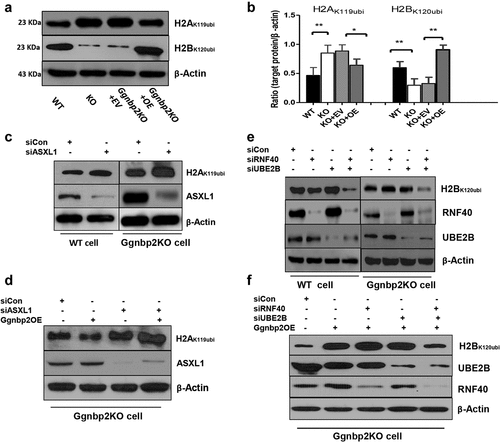
Next, we focused on how GGNBP2 regulates H2B ubiquitination. WT and Ggnbp2KO cells were treated with control scrambled siRNA, RNF40 siRNA and UBE2B siRNA. The results showed that the depletion of RNF40 or UBE2B separately did not decrease H2BK120ubiquitination (). Comparatively, when GC-2 cells were transfected with RNF40 siRNA together with UBE2B siRNA, the expression of H2BK120ub was significantly reduced (). To clarify whether GGNBP2 regulated H2B ubiquitination, Ggnbp2KO cells were transfected with UBE2B siRNA, RNF40 siRNA, and Ggnbp2 overexpression plasmids. As anticipated, RNF40 or UBE2B depletion did not impact H2Bk120ubiquitination (). We established that both RNF40 and UBE2B siRNA interference could inhibit H2B ubiquitination. In addition, we also found that Ggnbp2 overexpression increased siRNA-induced low levels of H2BK120 ubiquitination compared with GC-2 cells transfected with control siRNA.
GGNBP2 mediated H3K27 and H3K79 methylation by PRC2 subunits and histone H3K79 methyltransferase
Histone ubiquitination and histone H3 methylation’s crosstalk has been well documented. H2A ubiquitination represses gene expression via trans-histone crosstalk with H3K27trimethylation (H3K27me3) [Citation34], a histone marker linked to gene repression [Citation35]. Thus, we determined the expression of histone H3, K4, K9, K27 and K79 methylation and H3, K9, H4 and K16 acetylation in 18-day testis and GC-2 cells. Our results showed that the levels of histone H3K27me3 were significantly higher in both Ggnbp2KO testis and GC-2 cells, parallel with that of H2A ubiquitination, whereas the levels of histone H3K79me2 were significantly lower in both Ggnbp2KO testis and GC-2 cells, in accordance with H2B ubiquitination (). To verify this alteration, we transfected Ggnbp2KO GC-2 cells with overexpression plasmids. Our in vitro study showed that Ggnbp2 overexpression rescued the levels of H3K27me3 and H3K79me2 ().
Figure 6. GGNBP2 controlled patterns of histone modification.
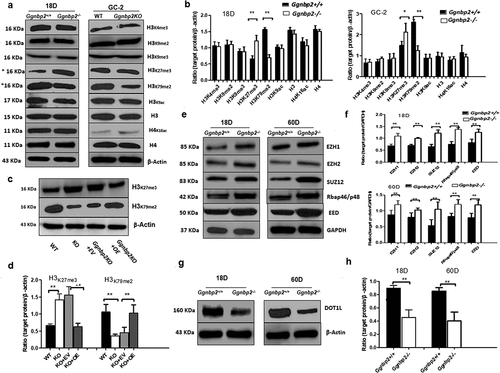
It has been reported that PRC2 catalyses the mono-, di- and tri-methylation of H3K27 [Citation36], while ubiquitylation of H2B was essential for subsequent trimethylation of H3K4 and H3K79 [Citation37]. H2BK120ubi stimulates H3K4 and H3K79 di- and tri-methylation in both budding yeast and human cells [Citation38], possibly through an H2Bub-methyltransferase bridge (Set1-COMPASS/Dot1). To further explore how histone ubiquitination affected methylation, we investigated the protein expression of PRC2 components and histone H3K79 methyltransferase DOT1L in juvenile and adult mouse testis. Three PcG proteins, enhancer of zeste 2 (EZH2), embryonic ectoderm development (EED) and suppressor of zeste 12 homolog (SUZ12) formed the core of PRC2 and mediated methylation on histone H3 lysine 27 (H3K27) [Citation39]. Histone-binding protein RBBP4 (or RbAp48/p46) and zinc finger protein AEBP2 bind to PRC2 and optimize its enzymatic activity in vitro [Citation40]. A recent study revealed that EZH1, the other sequence homologs of EZH, was part of a noncanonical PRC2 complex that catalyses the addition of methyl groups on H3K27 [Citation41]. Our results indicated that the expression of all the subunits of the PRC2 complex was increased in Ggnbp2 knockout total testis compared with WT, triggering the up-regulation of H3K27 methylation (). Simultaneously, GGNBP2-deficient testis showed a significant decrease in DOT1L expression (), explaining the downregulation of H3K79 di-methylation.
Discussion
Epigenetic changes involve histone proteins that function in the packaging of DNA and a series of DNA modifications [Citation42]. Nucleosomes comprise four core histone proteins, H2A, H2B, H3 and H4, wrapped by 147 base pairs of genomic DNA. They are further packaged into higher-order chromatin structures to compact the genomic DNA into the nucleus. Histones can change DNA-binding capacities and the interaction of other regulatory factors with DNA by conformational modification, resulting in alterations in gene activation [Citation43]. In response to DNA damage, cells undertake a series of responses, such as stopping transcription, inducing cell cycle arrest, repairing damaged DNA, and restoring normal cellular activities after repair [Citation44]. During this process, histone post-translation modification plays an essential role. The major ones are methylation, acetylation, phosphorylation, sumoylation, and ubiquitination [Citation45]. Such posttranslational modifications regulate the structure of chromatin and the recruitment of other proteins. Lukas et al. reported that histone H2AX phosphorylation (γ-H2AX) was induced by double-strand breaks (DSBs) and DNA replication stress and functioned as an important signal for initiating the downstream chromatin-associated events [Citation46]. In previous studies, we explored the function of γ-H2AX and staining pattern in Ggnbp2 knockout mouse testis and described that Ggnbp2 loss increased DNA DSB and led to DSB repair [Citation20]. Here, we confirmed that Ggnbp2 knockout increased both H2AK119 ubiquitination levels and decreased H2BK120 ubiquitination levels by protein–protein interaction. We also demonstrated trans-histone crosstalk between ubiquitination and methylation by regulating PRC2 subunits and methyltransferase DOT1L activity in Ggnbp2 knockout mice.
In the ubiquitination system, DUBs are responsible for reversing ubiquitination reactions by removing covalently attached ubiquitin molecules from substrates or polyubiquitinated chains and exerting a profound influence on many cellular pathway events, including DNA repair and damage response in mammalian cells. H2A ubiquitination, the major Ub conjugate in male meiosis, has been shown to play an important role during spermatogenesis and accumulates in the XY body and unsynapsed chromosomal regions undergoing meiotic silencing of unsynapsed chromatin (MSUC) [Citation32]. However, the absence of ubiquitinated H2A in the XY body does not impact XY body formation, sex chromosome inactivation, or meiotic progression, indicating that H2A ubiquitination is not essential for these processes [Citation47]. Here, we found that the staining of H2AK119ubi was mainly restricted to XY body in WT spermatocytes, whereas it was more diffused in Ggnbp2 knockout spermatocytes. Our findings showed that GGNBP2, ASXL1 and BAP1 could together form a complex. ASXL1 acted as a bridge to connect GGNBP2 and BAP1, with the latter functioning as DUB specific to H2AK119ubi. Without GGNBP2 interaction, the binding between ASXL1 and BAP1 would be disrupted. As a result, BAP1 could not enter the nucleus to deubiquitinate H2AK119, which led to an increase in the level of H2AK119ubi. To validate our in vivo results, in vitro si-RNA interference was performed. As expected, Ggnbp2 overexpression significantly rescued the H2AK119ubi level in mutant cells but did not restore the interaction of ASXL1 and BAP1, suggesting that BAP1 was in an inactivated state. Even though ASXL1 knockdown significantly increased H2AK119ubi expression in WT and mutant cells, BAP1 could not exert deubiquitinase activity in mutant cells transfected with Ggnbp2 overexpression plasmid. Together, these findings established GGNBP2 as an important negative regulator for H2AK119ubiquitination in DNA damage response.
Several studies have clarified the role of H2B ubiquitination in activating and silencing gene transcription [Citation48,Citation49]. Recent studies have begun focusing on the important role of H2Bubi-mediated nucleosome stabilization in maintaining the chromatin structure for the fidelity of transcriptional initiation. H2B ubiquitination can regulate initiation by stabilizing nucleosomes positioned over the promoters of repressed genes [Citation26]. Xu et al. showed that RNF20/Bre1p-mediated H2B ubiquitination regulated meiotic recombination by promoting chromatin relaxation and provided evidence that chloroquine, a chromatin relaxation agent, could treat some oligo or azoospermia patients with chromatin relaxation disorders [Citation28]. Protein structure analysis of recombinant nucleosomes indicated that H2B ubiquitination might be involved in the de-condensation of chromatin [Citation50]. In this study, we observed compromised spermatocyte chromatin compaction and nucleosome stability in Ggnbp2 knockout GC-2 cells in vitro and germ cells in vivo, which were more susceptible to MNase digestion and more soluble at very low salt concentrations. GGNBP2 might facilitate H2B ubiquitination, which promotes chromatin relaxation in spermatocytes. However, the mechanisms by which GGNBP2 regulates H2B ubiquitination levels remained undetermined. Our findings suggest that GGNBP2 interacts with UBE2B, a ubiquitin-conjugating E2, together with ubiquitin ligase E3 RNF40 to ubiquitinate H2BK120. Loss of GGNBP2 disconnects UBEB/RNF40 binding and affects H2B ubiquitination.
A key step in gene repression by Polycomb is the trimethylation of histone H3K27 by PCR2 to form H3K27me3, which provides a binding surface for PRC1 [Citation51]. Cao et al. demonstrated that the knockdown of PRC2 subunit SUZ12, which reduces Lys-27 H3 methylation by the EED-EZH2 complex, could reduce H2AK119ubi localization at silenced promoters [Citation52]. H2A ubiquitination seemed like a downstream regulator of Lys-27 H3 methylation. Our study demonstrated that H3K27me3 and PRC2 subunits EZH2, EZH, EED, SUE12 and Rbap46/p48 were up-regulated by Ggnbp2 depletion. In this study, we provided evidence showing that the regulation of GGNBP2 in histone H2A ubiquitination could be attributed to both the ubiquitin ligase and deubiquitinase. It has been established that both the PRC1 and PRC2 subunits are recruited to the damage site after DNA damage, suggesting a role for H2AK119ubi and H3K27me3 in transcription silencing in the damaged chromatin [Citation53]. Based on these findings, we concluded that Ggnbp2 loss increased DNA DSB, where more PRC2 subunits could be recruited, resulting in an up-regulation of H2AK119ubi and H3K27me3. In contrast to H2A ubiquitination and H3 methylation, H2B monoubiquitination is a prerequisite for H3K4 and H3K79 methylation. Further, this histone crosstalk has been confirmed to function unidirectionally [Citation37]. We also showed that H3K79me2 was down-regulated in Ggnbp2 knockout cells. In addition, a low level of the specific methyltransferase DOT1L expression was also observed in juvenile and adult testis. Thus, it can be hypothesized that GGNBP2 regulates H3K79 methylation by inhibiting histone methyltransferase DOT1L activity.
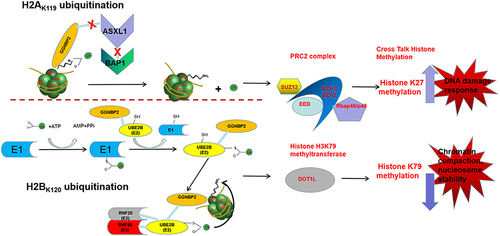
In conclusion, these results provided evidence suggesting the involvement of histone ubiquitination and methylation in the development of GGNBP2 loss resulting in azoospermia phenotype. We propose that Ggnbp2 knockout increased DNA damage response by promoting H2A ubiquitination and H3K27trimethylation, whereas it reduced nucleosome stability by decreasing H2B ubiquitination and H3K79dimethylation. Altogether, the present study shed new light on the mechanisms of the epigenetic phenomenon during spermatogenesis, such as the DNA damage repair process and its effects on sperm production problems (). Understanding the molecular mechanisms underlying Ggnbp2 deletion-related azoospermia and the role of GGNBP2 in regulating chromatin structure and gene expression could be useful in developing new diagnostic and therapeutic strategies for male infertility.
Figure 7. The mechanism of histone modifications regulated by GGNBP2.
Ethical approval
The animal studies were approved by the Animal Care and Use Committee of the First Hospital of Jilin University. All of the experimental procedures involving animals were conducted in accordance with the Institutional Animal Care guidelines of our hospital.
-) Clean Table S1.docx
Download MS Word (19.6 KB)supplement figure 1.tif
Download TIFF Image (2 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592294.2024.2381849
Additional information
Funding
References
- Ohbayashi T, Oikawa K, Iwata R, et al. Dioxin induces a novel nuclear factor, DIF-3, that is implicated in spermatogenesis. FEBS Lett. 2001;508(3):341–17. doi: 10.1016/s0014-5793(01)03039-3
- Lan ZJ, Hu Y, Zhang S, et al. GGNBP2 acts as a tumor suppressor by inhibiting estrogen receptor α activity in breast cancer cells. Breast Cancer Res Treat. 2016;158(2):263–276. doi: 10.1007/s10549-016-3880-2
- Li Y, Chen Z. Molecular cloning and characterization of LCRG1 a novel gene localized to the tumor suppressor locus D17S800-D17S930. Cancer Lett. 2004;209(1):75–85. doi: 10.1016/j.canlet.2003.11.034
- Yin F, Liu L, Liu X, et al. Downregulation of tumor suppressor gene ribonuclease T2 and gametogenetin binding protein 2 is associated with drug resistance in ovarian cancer. Oncol Rep. 2014;32(1):362–372. doi: 10.3892/or.2014.3175
- Zhu Z, Lou C, Zheng Z, et al. ZFP403, a novel tumor suppressor, inhibits the proliferation and metastasis in ovarian cancer. Gynecol Oncol. 2017;147(2):418–425. doi: 10.1016/j.ygyno.2017.08.025
- Zhan A, Lei B, Wu H, et al. GGNBP2 suppresses the proliferation, invasion, and migration of human glioma cells. Oncol Res. 2017;25(5):831–842. doi: 10.3727/096504016x14816726393937
- Yang Z, Wang Y, Ma L. Effects of gametogenetin-binding protein 2 on proliferation, invasion and migration of prostate cancer PC-3 cells. Andrologia. 2020;52(2):e13488. doi: 10.1111/and.13488
- Chen A, Li J, Song L, et al. GGNBP2 is necessary for testis morphology and sperm development. Sci Rep. 2017;7(1):2998. doi: 10.1038/s41598-017-03193-y
- Liu L, He Y, Guo K, et al. Ggnbp2-null mutation in mice leads to male infertility due to a defect at the spermiogenesis stage. Am J Pathol. 2017;187(11):2508–2519. doi: 10.1016/j.ajpath.2017.07.016
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14(11):1008–1016. doi: 10.1038/nsmb1337
- Chen K, Zeng J, Sun Y, et al. Junction plakoglobin regulates and destabilizes HIF2α to inhibit tumorigenesis of renal cell carcinoma. Cancer Commun. 2021;41(4):316–332. doi: 10.1002/cac2.12142
- Tamburri S, Conway E, Pasini D. Polycomb-dependent histone H2A ubiquitination links developmental disorders with cancer. Trends Genet. 2022;38(4):333–352. doi: 10.1016/j.tig.2021.07.011
- Mattiroli F, Penengo L. Histone ubiquitination: an integrative signaling platform in genome stability. Trends Genet. 2021;37(6):566–581. doi: 10.1016/j.tig.2020.12.005
- Wang H, Wang L, Erdjument-Bromage H, et al. Role of histone H2A ubiquitination in polycomb silencing. Nature. 2004;431(7010):873–878. doi: 10.1038/nature02985
- Morreale FE, Walden H. Types of ubiquitin ligases. Cell. 2016;165(1):248–.e1. doi: 10.1016/j.cell.2016.03.003
- Guo Y, Zhang H, Yao L, et al. Systematic analysis of the ubiquitome in mouse testis. Proteomics. 2021;21(15):e2100025. doi: 10.1002/pmic.202100025
- Korenfeld HT, Avram-Shperling A, Zukerman Y, et al. Reversal of histone H2B mono-ubiquitination is required for replication stress recovery. DNA Repair (Amst). 2022;119(103387):103387. doi: 10.1016/j.dnarep.2022.103387
- Feng L, Wang J, Chen J. The Lys63-specific deubiquitinating enzyme BRCC36 is regulated by two scaffold proteins localizing in different subcellular compartments. J Biol Chem. 2010;285(40):30982–30988. doi: 10.1074/jbc.M110.135392
- Ng HM, Wei L, Lan L, et al. The Lys63-deubiquitylating enzyme BRCC36 limits DNA break processing and repair. J Biol Chem. 2016;291(31):16197–16207. doi: 10.1074/jbc.M116.731927
- Guo K, He Y, Liu L, et al. Ablation of Ggnbp2 impairs meiotic DNA double-strand break repair during spermatogenesis in mice. J Cell Mol Med. 2018;22(10):4863–4874. doi: 10.1111/jcmm.13751
- Sowa ME, Bennett EJ, Gygi SP, et al. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138(2):389–403. doi: 10.1016/j.cell.2009.04.042
- Koken MH, Reynolds P, Jaspers-Dekker I, et al. Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc Natl Acad Sci USA. 1991;88(20):8865–8869. doi: 10.1073/pnas.88.20.8865
- Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Sci (New Y NY). 2000;287(5452):501–504. doi: 10.1126/science.287.5452.501
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, et al. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11(1):261–266. doi: 10.1016/s1097-2765(02)00826-2
- Suryavathi V, Khattri A, Gopal K, et al. Novel variants in UBE2B gene and idiopathic male infertility. J Androl. 2008;29(5):564–571. doi: 10.2164/jandrol.107.004580
- Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci USA. 2009;106(39):16686–16691. doi: 10.1073/pnas.0907862106
- Zhang X, Xiao Z, Chen Z, et al. Comparative proteomics analysis of the proteins associated with laryngeal carcinoma-related gene 1. Laryngoscope. 2006;116(2):224–230. doi: 10.1097/01.mlg.0000191470.71454.a1
- Xu Z, Song Z, Li G, et al. H2B ubiquitination regulates meiotic recombination by promoting chromatin relaxation. Nucleic Acids Res. 2016;44(20):9681–9697. doi: 10.1093/nar/gkw652
- Li JP, Guo WB, He JC, et al. Establishing a mouse model of sertoli-cell-only syndrome by administration of busulfan. Zhonghua Nan Ke Xue. 2013;19(4):300–305.
- Dia F, Strange T, Liang J, et al. Preparation of meiotic chromosome spreads from mouse spermatocytes. J Vis Exp. 2017129. doi: 10.3791/55378
- Wang T, Gao H, Li W, et al. Essential role of histone replacement and modifications in male fertility. Front Genet. 2019;10(962). doi: 10.3389/fgene.2019.00962
- Baarends WM, Wassenaar E, van der Laan R, et al. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol Cell Biol. 2005;25(3):1041–1053. doi: 10.1128/mcb.25.3.1041-1053.2005
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, et al. Histone H2A deubiquitinase activity of the polycomb repressive complex PR-DUB. Nature. 2010;465(7295):243–247. doi: 10.1038/nature08966
- de Ayala Alonso A G, Gutiérrez L, Fritsch C, et al. A genetic screen identifies novel polycomb group genes in drosophila. Genetics. 2007;176(4):2099–2108. doi: 10.1534/genetics.107.075739
- Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009
- Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi: 10.1038/nature09784
- Briggs SD, Xiao T, Sun ZW, et al. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418(6897):498. doi: 10.1038/nature00970
- Kim J, Guermah M, McGinty RK, et al. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137(3):459–471. doi: 10.1016/j.cell.2009.02.027
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, et al. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of zeste protein. Genes Devel. 2002;16(22):2893–2905. doi: 10.1101/gad.1035902
- Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15(1):57–67. doi: 10.1016/j.molcel.2004.06.020
- Shen X, Liu Y, Hsu YJ, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32(4):491–502. doi: 10.1016/j.molcel.2008.10.016
- Wang X, Zhang Q, Cao X. Reversing epigenetic repression of transposable elements for improving tumor immunogenicity. Cancer Commun. 2022;42(3):266–268. doi: 10.1002/cac2.12261
- Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461(7261):193–198. doi: 10.1038/nature08450
- Sirbu BM, Cortez D. DNA damage response: three levels of DNA repair regulation. Cold Spring Harb Perspect Biol. 2013;5(8):a012724. doi: 10.1101/cshperspect.a012724
- Nottke A, Colaiácovo MP, Shi Y. Developmental roles of the histone lysine demethylases. Development (Cambridge, England). Development. 2009;136(6):879–889. doi: 10.1242/dev.020966
- Lukas J, Lukas C, Bartek J. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol. 2011;13(10):1161–1169. doi: 10.1038/ncb2344
- Szczepanski AP, Wang L. Emerging multifaceted roles of BAP1 complexes in biological processes. Cell Death Discov. 2021;7(1):20. doi: 10.1038/s41420-021-00406-2
- Pavri R, Zhu B, Li G, et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125(4):703–717. doi: 10.1016/j.cell.2006.04.029
- Gardner RG, Nelson ZW, Gottschling DE. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol Cell Biol. 2005;25(14):6123–6139. doi: 10.1128/mcb.25.14.6123-6139.2005
- Yamashita K, Shinohara M, Shinohara A. Rad6-Bre1-mediated histone H2B ubiquitylation modulates the formation of double-strand breaks during meiosis. Proc Natl Acad Sci USA. 2004;101(31):11380–11385. doi: 10.1073/pnas.0400078101
- Kalb R, Latwiel S, Baymaz HI, et al. Histone H2A monoubiquitination promotes histone H3 methylation in polycomb repression. Nat Struct Mol Biol. 2014;21(6):569–571. doi: 10.1038/nsmb.2833
- Cao R, Tsukada Y, Zhang Y. Role of bmi-1 and Ring1A in H2A ubiquitylation and hox gene silencing. Mol Cell. 2005;20(6):845–854. doi: 10.1016/j.molcel.2005.12.002
- Sekiguchi M, Matsushita N. DNA damage response regulation by histone ubiquitination. Int J Mol Sci. 2022;23(15):8187. doi: 10.3390/ijms23158187
