Abstract
Transcriptional feedback loops in Arabidopsis circadian clock is composed of more repressive components, while the knowledge of activation mechanism remains limited. We recently reported 2 members from a family of NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED genes, LNK1 and LNK2, dynamically interact with morning-phased transcriptional factors, like CIRCADIAN CLOCK ASSOCIATED1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), REVEILLE8 (RVE8) and RVE4, and function as coactivators for the expression of TIMING OF CAB EXPRESSION1 (TOC1) and PSEUDO-RESPONSE REGULATOR5 (PRR5) via transcriptional factors RVE8 and RVE4. Here we provide evidence that both LNK1 and LNK2 play critical role in the transcriptional activation of PRR5, LNK1 may contribute more than LNK2 did under experimental conditions. We also identified that both LNK1 and LNK2 recruitment to the evening element of PRR5 promoter via LNK1-RVE8 or LNK2-RVE8 proteins complex through electrophoretic mobility shift assay. Therefore LNK1 and LNK2 function as coactivator of dawn-phased MYB-like transcription factors, such as RVE8 in morning complex to regulate the target genes expression.
Living organisms take fitness advantage of circadian control and coordinate with the daily and seasonal changes in environments.Citation1-4 On the basis of complicated diurnal and nocturnal growth and metabolism, the Arabidopsis clock consists of multiple transcriptional and translational feedback loops to contribute the rhythmic outputs.Citation5,6 The circadian components, like PSEUDO-RESPONSE REGULATORs, TOC1/PRR5/PRR7/PRR9 and CIRCADIAN CLOCK ASSOCIATED1 (CCA1)/ LATE ELONGATED HYPOCOTYL (LHY), require co-repressors DE-ETIOLATED1 (DET1), TOPLESS/TOPLESS-RELATED (TPL/TPR) or CCA1 HIKING EXPEDITION (CHE), forming repressive complex for the transcriptional repression of multiple genes.Citation7-9 The REVEILLEs, RVE8, RVE6 and RVE4, as antagonists of CCA1, LHY to a certain degree, perform as positive regulators in the feedback loops.Citation10,11 The mechanism by which Myb domain transcription factors promote the expression of PRR5, TOC1, PRR9, PRR7, LUX ARRHYTHMO (LUX), EARLY FLOWERING 4 (ELF4), and GIGANTEA (GI) is largely unknown. NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED family was first identified as light strongly induced genes through a genomewide transcriptome analysis. TOC1 binds to the promoter of LNK1 and LNK2 as a repressor, LNK1 and LNK2 directly or indirectly promote the expression of PRR5 and ELF4.Citation12 Recently we reported that LNK1 and LNK2 dynamically interact with multiple dawn-phased transcriptional factors, including CCA1, LHY, RVE8 and RVE4; LNK1 is demonstrated to function as coactivator of RVE8 for the expression of PRR5 and TOC1.Citation13
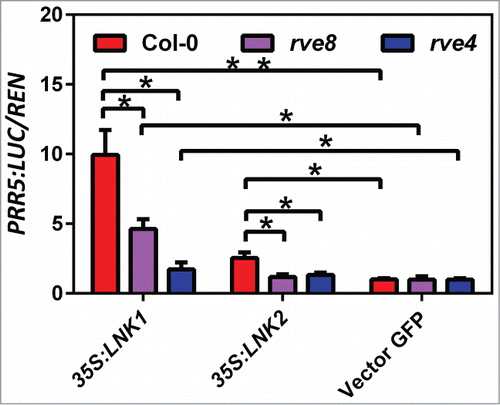
In addition to the red light signaling input to the clock, both LNK1 and LNK2 play similar role in phototmorphogenesis and flowering time control. LNK2, like LNK1, highly associated with the morning clock components CCA1 and LHY. Together with LNK1, LNK2 behaved transcriptional activity in yeast assay of AD/LNK-BD. To further determine the individual contributions of LNK1 and LNK2 in the circadian core oscillator, we identified and compared their ability to promote the expression of PRR5 via transcriptional factors RVE4 and RVE8. In Col-0, rve8 or rve4 mutant protoplasts carrying PRR5:LUC reporter, the transiently over-expressed 35S:LNK1-FLAG or 35S:LNK2-FLAG significantly increased PRR5 expression at Zeitgeber time 6 (ZT6). In addition, We observed much greater PRR5:LUC activity in Col-0, rve8 and rve4 mutant with LNK1 compared with LNK2 (). This indicates that LNK1 could be more critical than LNK2 in the promotion of PRR5 transcription via RVE8 or RVE4 under experimental conditions. Interestingly, we also noted that the activation of PRR5:LUC expression by LNK2 was distinctly blocked in rve8 or rve4 mutant compared with in Col-0, which possibly due to the activation ability of LNK2 is not as strong as LNK1. In summary, our results suggest that the activation of PRR5 by RVE8 or RVE4 requires LNK1 and LNK2 ().
Figure 1. The transcriptional activation of PRR5 by RVE8 and RVE4 requires LNK1 and LNK2. Relative expression of PRR5:LUC/REN (35S:RenLUC) normalized to Vector GFP control, in transiently transfected Arabidopsis protoplasts constitutively overexpressing LNK1 or LNK2 in Col-0 and in rve8 and rve4 mutant backgrounds (sampled at ZT6). Horizontal lines indicate values that are significantly different (ANOVA; *P < 0.05, **P < 0.01).
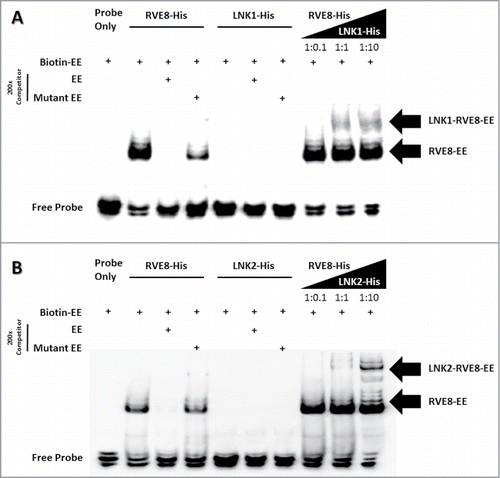
As noted before, LNK1 does not possess the DNA-binding ability when we performed the EMSA assay with EE element in vitro. LNK1 or LNK2 physically interact with multiple clock core transcriptional factors including CCA1, LHY, RVE4 and RVE8.Citation13 To investigate whether LNK1 or LNK2 serve as coactivators through forming morning protein complex with circadian transcription factors, we purified Escherichia coli-produced recombinant His-tagged LNK1, LNK2 and RVE8 to performed electrophoretic mobility shift assay (EMSA). RVE8-His was able to directly bind biotin-labeled evening element (EE) probe alone in vitro. With addition of LNK1-His or LNK2-His, extra-large potential RVE8-LNK1-EE and RVE8-LNK2-EE protein complex were detected along with gradient increased LNKs (). Therefore, our results are consistent with the function for LNK1 and LNK2 as coactivator of RVE8 within the core circadian clock.
Figure 2. Recruitment of LNK1 and LNK2 to EE probe via interaction with RVE8. EMSA of a probe including the EE with RVE8-His and RVE8-His plus LNK1-His or LNK2-His which purified from E. coli BL21 (DE3). The arrow indicates the RVE8/EE, LNK1/RVE8/EE and LNK2/RVE8/EE complex.
Taken together, in vivo transcriptional assay in protoplast presented here indicates that LNK1 play more significant role than LNK2 in the promotion of target genes expression by RVE8 and RVE4. On the basis of EMSA assay, LNK1 could not bind directly to the EE-containing element. Activation activity of LNK1 needs morning-phase Myb-like transcriptional factors, such as RVE8 or RVE4, through proteins physically interaction. LNK1 was recruited to EE of promoter of PRR5 and TOC1 for further activation.Citation13 Here we detected larger potential RVE8-LNK1-EE and RVE8-LNK2-EE complex by EMSA assay in vitro, which support the hypothesis that formation of putative morning complex (MC), like LNK1-RVE8 and LNK2-RVE8 to contribute the new regulatory mechanism of circadian clock in the morning. It is known that in the early evening, the circadian regulated evening complex (EC) which consists of protein ELF3, ELF4 and LUX, function as repressors for the PIF4, PIF5 expression.Citation14 Furthermore, LUX could also recruit ELF3 to the PRR9 promoter region.Citation15 Considering the interaction of LNK1, LNK2 with CCA1, LHY, RVE8 and RVE4, LNKs may serve with specific transcriptional factors with favorably phase. Therefore, we propose that LNK1, LNK2, and by extension with other LNK family members, serves as morning complex transcriptional components in the Arabidopsis circadian clock to benefit plant growth and development.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Jun-Xian He, Bingchun Zhao and Rui Li for technical help.
Funding
This work was supported by grants from the Special Program for Key Basic Research of the Ministry of Science and Technology of China (2012CB126303), National Science Foundation of China (31071247), Program for New Century Excellent Talents in University of Ministry of Education of China (NCET-13-0771), Hebei Science Fund for Distinguished Young Scholars (C2011205034, CPRC036, 20100502, GCC2014063) to X.X and funds for Creative Research Groups of Hebei Province, China (LJRC025).
References
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 2005; 309:630-3; PMID:16040710; http://dx.doi.org/10.1126/science.1115581
- Wang W, Barnaby JY, Tada Y, Li H, Tor M, Caldelari D, et al. Timing of plant immune responses by a central circadian regulator. Nature 2011; 470:110-4; PMID:21293378; http://dx.doi.org/10.1038/nature09766
- Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 2009; 457:327-31; PMID:19029881; http://dx.doi.org/10.1038/nature07523
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 2008; 9:R130; PMID:18710561; http://dx.doi.org/10.1186/gb-2008-9-8-r130
- McClung CR. Wheels within wheels: new transcriptional feedback loops in the Arabidopsis circadian clock. F1000Prime Rep 2014; 6:2; PMID:24592314; http://dx.doi.org/10.12703/P6-2
- Hsu PY, Harmer SL. Wheels within wheels: the plant circadian system. Trends Plant Sci 2014; 19:240-9; PMID:24373845; http://dx.doi.org/10.1016/j.tplants.2013.11.007
- Lau OS, Huang X, Charron JB, Lee JH, Li G, Deng XW. Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol Cell 2011; 43:703-12; PMID:21884973; http://dx.doi.org/10.1016/j.molcel.2011.07.013
- Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 2009; 323:1481-5; PMID:19286557; http://dx.doi.org/10.1126/science.1167206
- Wang L, Kim J, Somers DE. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci 2013; 110:761-6; PMID:23267111; http://dx.doi.org/10.1073/pnas.1215010110
- Hsu PY, Devisetty UK, Harmer SL. Accurate timekeeping is controlled by a cycling activator in Arabidopsis. Elife 2013; 2:e00473; PMID:23638299; http://dx.doi.org/10.7554/eLife.00473
- Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, et al. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the arabidopsis circadian clock. PLoS Genet 2011; 7:e1001350; PMID:21483796; http://dx.doi.org/10.1371/journal.pgen.1001350
- Rugnone ML, Faigon Soverna A, Sanchez SE, Schlaen RG, Hernando CE, Seymour DK, et al. LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc Natl Acad Sci U S A 2013; 110:12120-5; PMID:23818596; http://dx.doi.org/10.1073/pnas.1302170110
- Xie Q, Wang P, Liu X, Yuan L, Wang L, Zhang C, et al. LNK1 and LNK2 are transcriptional coactivators in the arabidopsis circadian oscillator. Plant Cell 2014; 26:2843-57; PMID:25012192; http://dx.doi.org/10.1105/tpc.114.126573
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 2011; 475:398-402; PMID:21753751; http://dx.doi.org/10.1038/nature10182
- Chow BY, Helfer A, Nusinow DA, Kay SA. ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signal Behav 2012; 7:170-3; PMID:22307044; http://dx.doi.org/10.4161/psb.18766
