Abstract
Protein intrinsic disorder (ID), referring to the lack of a fixed tertiary structure, is significant in signaling and transcription. We recently characterized ID in 6 phylogenetically representative Arabidopsis thaliana NAC transcription factors. Their transcription regulatory domains are mostly disordered but contain short, functionally important regions with structure propensities known as molecular recognition features. Here, we analyze for NAC subgroup-specific ID patterns. Some subgroups, such as the VND subgroup implicated in secondary cell wall biosynthesis, and the NAP/SHYG subgroup have highly conserved ID profiles. For the stress-associated ATAF1 subgroup and the CUC/ORE1 subgroup involved in development, only sub clades have similar ID patterns. For similar ID profiles, conserved molecular recognition features and sequence motifs represent likely functional determinants of e.g. transcriptional activation and interactions. Based on our analysis, we suggest that ID profiling of regulatory proteins in general can be used to guide identification of interaction partners of network proteins.
Proteins with intrinsic disorder (ID) lack a defined tertiary structure but nonetheless play important biological roles. Upon interaction, ID regions may fold,Citation1 but interactions may also take place through for example molecular recognition features (MoRFs), which form local structure upon binding.Citation2 Conserved sequence motifs, linear motifs, often form the core of interactions sites in ID regions.Citation3
Proteins with ID are over represented in signaling and transcription which involve hubs with multiple protein partners functioning in regulatory networks.Citation4,5 One key plant-specific signaling hub is Radical Induced Cell Death1 (RCD1), which interacts with several transcription factors associated with stress responses.Citation6 The promiscuity of RCD1 was explained by the flexibility associated with transcription factor ID.Citation7 The plant-specific NAM, ATAF, CUC (NAC) transcription factors play central roles in development and stress-responses.Citation8 They consist of a conserved DNA-binding domain, the NAC domain, and varying disordered C-terminal transcription regulatory domains (TRDs) () with interspersed conserved sequence motifs.Citation9 Our recent workCitation10 characterizing NAC ID suggested that the large disordered TRD of ANAC0046 forms an entropic chain which is well-suited for molecular fishing of e.g., RCD1 ().
Figure 1. ID in NAC transcription factors. (A) Schematic domain structure of a typical NAC transcription factor. The N-terminal DNA-binding NAC domain (green) forms a twisted β sheet (Protein Data Bank accession 1UT7) followed by a disordered C-terminus (different conformers in gray) encompassing transcription regulatory activity and a large interaction potential.Citation10 (B) Predicted disorder plotted as a function of alignment position for representative NAC proteins. Alignment position was determined from a multiple alignment of complete NAC proteins generated by ClustalX using MEGA4 softwareCitation28 followed by manual adjustments. The disorder tendency was predicted using metaPrDOS,Citation11 which integrates predictions from 5 different prediction methods. The threshold for prediction of ID is 0.5. The position of the NAC domain is shown by a green bar, while the position of the disordered transcription regulatory domain is shown by a gray bar.
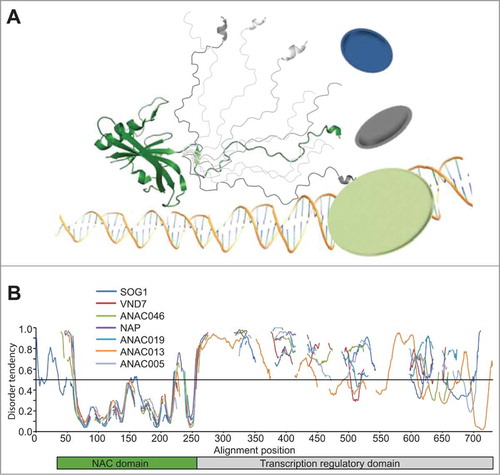
Here, we expand the study of NAC ID to the subgroup-level. Due to the lack of sequence similarity of the TRDs previous phylogenetic NAC analysis was based on only the NAC domain.Citation9 In this study, complete NAC sequences were used to derive dendograms for all Arabidopsis NAC proteins. ID predictions using metaPrDOSCitation11 confirmed that the ID patterns are not conserved in the NAC family ().Citation9 However, conserved ID patterns were reveled for several NAC subgroups (), in accordance with the suggestion that disorder patterns are constrained compared to sequences.Citation12,13
Figure 2. Phylogenetic NAC subgroups and predicted ID in NAC transcription regulatory domains. (Left) Phylogentic NAC subgroups derived from a tree generated using MEGA4 software based on a multiple alignment of complete Arabidopsis NAC proteins.Citation9 The color underlining the protein names corresponds to the color of the graphs shown at right. (Right) Predicted disorder as a function of alignment position for the transcription regulatory domains of NAC proteins from the subgroups shown. Alignment position and disorder tendency were determined as described in . The NAC domains were defined based on the multiple alignments and removed from the representation. Gaps in the resulting alignments were manually adjusted. The first amino acid residue of the transcription regulatory domain was defined as alignment position one. The approximate position of MoRFs predicted using Biomine MoRFpredCitation29 and sequence motifs identified using the MEME suite,Citation30 present in half or more of the sequences, is indicated by M or a colored bar, respectively. The letters in the bars refer to a previously identified sequence motif.Citation9 In addition, 2 novel sequence motifs, designated EF and v and having the consensus sequences E[KE][ED][DEM][YF][IL]E[MI][ND]DL and [RIT][DH]SLIP[LPQ][LTV][NV][NS], respectively, were identified in this study. The NAC proteins included in the analysis were named as previously suggested.Citation9 In addition, recently introduced acronyms refer to the following nomenclature: SMB, ANAC033, At1g79580; BRN1, ANAC015, At1g33280; BRN2, ANAC070, At4g10350; SHYG, ANAC047, At3g04070.
![Figure 2. Phylogenetic NAC subgroups and predicted ID in NAC transcription regulatory domains. (Left) Phylogentic NAC subgroups derived from a tree generated using MEGA4 software based on a multiple alignment of complete Arabidopsis NAC proteins.Citation9 The color underlining the protein names corresponds to the color of the graphs shown at right. (Right) Predicted disorder as a function of alignment position for the transcription regulatory domains of NAC proteins from the subgroups shown. Alignment position and disorder tendency were determined as described in Figure 1 . The NAC domains were defined based on the multiple alignments and removed from the representation. Gaps in the resulting alignments were manually adjusted. The first amino acid residue of the transcription regulatory domain was defined as alignment position one. The approximate position of MoRFs predicted using Biomine MoRFpredCitation29 and sequence motifs identified using the MEME suite,Citation30 present in half or more of the sequences, is indicated by M or a colored bar, respectively. The letters in the bars refer to a previously identified sequence motif.Citation9 In addition, 2 novel sequence motifs, designated EF and v and having the consensus sequences E[KE][ED][DEM][YF][IL]E[MI][ND]DL and [RIT][DH]SLIP[LPQ][LTV][NV][NS], respectively, were identified in this study. The NAC proteins included in the analysis were named as previously suggested.Citation9 In addition, recently introduced acronyms refer to the following nomenclature: SMB, ANAC033, At1g79580; BRN1, ANAC015, At1g33280; BRN2, ANAC070, At4g10350; SHYG, ANAC047, At3g04070.](/cms/asset/74914a33-0dc6-48fa-83fb-90b066a076ee/kpsb_a_1010967_f0002_oc.jpg)
The large subgroup II-1, implicated in secondary cell wall biosynthesis,Citation14 showed high conservation of ID patterns (). The ID profiles of the TRDs have 3 significant dips at positions which generally coincide with a MoRF, suggestive of local structure. Two of these regions coincide with previously defined sequence motifs, the LP and WQ motifs.Citation9,15 The WQ motif is necessary for transcriptional activity of SND,Citation1,15 whereas the function of the LP motif remains elusive. However, conservation of this motif and coincidence with a MoRF strongly suggests functional significance. The results obtained for subgroup II-1 demonstrate that functional determinants can be revealed by in silico analysis of ID regions.
For subgroup II-3, no common disorder profile was identified (), which may reflect functional diversity of this subgroup. However, further division revealed a common profile for the clades containing senescence associated NAC proteins such as ORE1Citation16 and ANAC046.Citation10 In addition to their common ID pattern, these proteins contain a C-terminal MoRF, which is a functional hotspot in ANAC046 mediating both interaction with RCD1 and transcriptional activity ().Citation10 Cleary, the corresponding MoRF region in the other NAC proteins represents a likely interaction determinant. A common ID pattern is not prevalent for the other subgroup II-3 clades (), containing the CUC proteins, which play significant roles in development.Citation17 This may reflect specificity of their molecular interactions.
Subgroup III-2 also contains biologically significant NAC proteins, e.g. NAP which is a positive regulator of senescence,Citation18 and SHYG which promotes protection against drowning.Citation19 Members of this subgroup display significant ID immediately following the NAC domain and a decrease in the ID propensity toward the C-termini, which encompass both a predicted MoRF and sequence motif ()Citation9 suggestive of a functional hotspot. Subclade-based division of stress-associated subgroup III-3, known as ATAF1, is needed to reveal common ID profiles (). ANAC019/055/072, with functional redundancy in abiotic stress responses,Citation20 share a significant dip in the disorder profile. The rest of the subgroup III-3 NAC proteins also have similar ID patterns with putative common interaction determinants. Similarities of the predicted ID profiles were also revealed for the subgroup VII-2 NAC proteins (), which remain to be characterized. Two of the 5 identified sequence motifs, L and f,Citation9 map to regions with low ID propensity, and one of these coincides with a MoRF. The tree subgroup IV-2 members (), ANAC013/016/017, also share ID features. These NAC proteins function in oxidative stress responses.Citation21–Citation23 and ANAC0013 and ANAC017 may mediate crosstalk between oxidative stress-responsive signaling pathways and mitochondrial retrograde regulation,Citation21,23 whereas ANAC016 may connect stress-signaling and senescence.Citation22 Whether the newly identified conserved sequence motif (EF)/MoRF is implicated in a common interaction and biological function remains to be studied. Like the rest of the NAC proteins (), the subgroup IV-2 NAC proteins contain a long ID region at the N-terminus of the TRD, which is likely to be a malleable linker to the NAC domain. Due to their transmembrane region (TM), ANAC013/016/017 were originally classified as NTM1-Like (NTL) NAC proteins. For the other NTL subgroups (I-1,4; IV-1,2 VII-1), common ID profiles were not identified. This was also the case for subgroup IX-1 (). Members of this subgroup contain an N-terminal extension of the NAC domain () and have diverse functions in e.g., responses to DNA damage (SOG1)Citation24 and secondary cell wall development (SND1/2).Citation25 This may explain the lack of common ID features.
Although the NAC TRD sequences have diverged greatly, probably reflecting the generally fast evolution of ID regions,Citation26,27 similar ID patterns can be distinguished for several NAC subgroups suggestive of conserved disorder-related functions. Further analysis will reveal if conserved ID profiles reflect common interaction partners or if adaptive mutations, allowing different interaction partners, are hidden in the common ID-features. The results obtained here and in a previous study of the Myc transcription factorsCitation12 suggest that ID predictions may eventually complement multiple alignments as a starting point for construction of dendrograms and thereby for dissection of evolutionary relationships. Importantly, since ID proteins constitute a significant part of regulatory networks and have an enormous interaction potential,Citation5 in silico analysis of ID regions, as presented here, should be used to direct identification of novel components of regulatory network, an important task in plant biology.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 2005; 6:197-08; PMID:15738986; http://dx.doi.org/10.1038/nrm1589
- Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, Dunker AK. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res 2007; 6:2351-66; PMID:17488107; http://dx.doi.org/10.1021/pr0701411
- Gould CM, Diella F, Via A, Puntervoll P, Gemund C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne JC, Chica C, et al. ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res 2010; 38:D167-80; PMID:19920119; http://dx.doi.org/10.1093/nar/gkp1016
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 2004; 337:635-45; PMID:15019783; http://dx.doi.org/10.1016/j.jmb.2004.02.002
- Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J 2005; 272:5129-48; PMID:16218947; http://dx.doi.org/10.1111/j.1742-4658.2005.04948.x
- Jaspers P, Blomster T, Brosche M, Salojarvi J, Ahlfors R, Vainonen JP, Reddy RA, Immink R, Angenent G, Turck F, et al. Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J 2009; 60:268-79; PMID:19548978; http://dx.doi.org/10.1111/j.1365-313X.2009.03951.x
- Kragelund BB, Jensen MK, Skriver K. Order by disorder in plant signaling. Trends Plant Sci 2012; 17:625-32; PMID:22819467; http://dx.doi.org/10.1016/j.tplants.2012.06.010
- Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 2005; 10:79-87; PMID:15708345; http://dx.doi.org/10.1016/j.tplants.2004.12.010
- Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O'Shea C, Skriver K. The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem J 2010; 426:183-96; PMID:19995345; http://dx.doi.org/10.1042/BJ20091234
- O'Shea C, Kryger M, Stender EG, Kragelund BB, Willemoes M, Skriver K. Protein intrinsic disorder in Arabidopsis NAC transcription factors: Transcriptional activation by ANAC013 and ANAC046 and their interactions with Radical Induced Cell Death1. Biochem J 2015; 465:281-94; PMID:25348421; http://dx.doi.org/10.1042/BJ20141045
- Ishida T, Kinoshita K. Prediction of disordered regions in proteins based on the meta approach. Bioinformatics 2008; 24:1344-8; PMID:18426805; http://dx.doi.org/10.1093/bioinformatics/btn195
- Mahani A, Henriksson J, Wright AP. Origins of Myc proteins–using intrinsic protein disorder to trace distant relatives. PLoS One 2013; 8:e75057; PMID:24086436; http://dx.doi.org/10.1371/journal.pone.0075057
- Toth-Petroczy A, Oldfield CJ, Simon I, Takagi Y, Dunker AK, Uversky VN, Fuxreiter, M. Malleable machines in transcription regulation: the mediator complex. PLoS Comput Biol 2008; 4:e1000243; PMID:19096501; http://dx.doi.org/10.1371/journal.pcbi.1000243
- Zhong R, Lee C, Ye ZH. Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol Plant 2010; 3:1087-103; PMID:20935069; http://dx.doi.org/10.1093/mp/ssq062
- Ko JH, Yang SH, Park AH, Lerouxel O, Han KH. ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J 2007; 50:1035-48; PMID:17565617; http://dx.doi.org/10.1111/j.1365-313X.2007.03109.x
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Köhler B, Mueller-Roeber B. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 2010; 62:250-64; PMID:20113437; http://dx.doi.org/10.1111/j.1365-313X.2010.04151.x
- Zhong R, Lee C, Ye ZH. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci 2010; 15:625-32; PMID:20833576; http://dx.doi.org/10.1016/j.tplants.2010.08.007
- Guo Y, Gan S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 2006; 46:601-12; PMID:16640597; http://dx.doi.org/10.1111/j.1365-313X.2006.02723.x
- Rauf M, Arif M, Fisahn J, Xue GP, Balazadeh S, Mueller-Roeber B. NAC transcription factor speedy hyponastic growth regulates flooding-induced leaf movement in Arabidopsis. Plant Cell 2013; 25:4941-55; PMID:24363315; http://dx.doi.org/10.1105/tpc.113.117861
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004; 16:2481-98; PMID:15319476; http://dx.doi.org/10.1105/tpc.104.022699
- De CI, Vermeirssen V, Van AO, Vandepoele K, Murcha MW, Law SR, Inzé A, Ng S, Ivanova A, Rombaut D, et al. The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 2013; 25:3472-90; PMID:24045019; http://dx.doi.org/10.1105/tpc.113.117168
- Kim YS, Sakuraba Y, Han SH, Yoo SC, Paek NC. Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant Cell Physiol 2013; 54:1660-72; PMID:23926065; http://dx.doi.org/10.1093/pcp/pct113
- Ng S, Ivanova A, Duncan O, Law SR, Van AO, De Clercq I, Wang Y, Carrie C, Xu L, Kmiec B, et al. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 2013; 25:3450-71; PMID:24045017; http://dx.doi.org/10.1105/tpc.113.113985
- Yoshiyama K, Conklin PA, Huefner ND, Britt AB. Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc Natl Acad Sci U S A 2009; 106:12843-8; PMID:19549833; http://dx.doi.org/10.1073/pnas.0810304106
- Hussey SG, Mizrachi E, Spokevicius AV, Bossinger G, Berger DK, Myburg AA. SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol 2011; 11:173; PMID:22133261; http://dx.doi.org/10.1186/1471-2229-11-173
- Brown CJ, Johnson AK, Daughdrill GW. Comparing models of evolution for ordered and disordered proteins. Mol Biol Evol 2010; 27:609-21; PMID:19923193; http://dx.doi.org/10.1093/molbev/msp277
- Brown CJ, Johnson AK, Dunker AK, Daughdrill GW. Evolution and disorder. Curr Opin Struct Biol 2011; 21:441-6; PMID:21482101; http://dx.doi.org/10.1016/j.sbi.2011.02.005
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24:1596-9; PMID:17488738; http://dx.doi.org/10.1093/molbev/msm092
- Disfani FM, Hsu WL, Mizianty MJ, Oldfield CJ, Xue B, Dunker AK, Uversky, VN, Kurgan, L. MoRFpred, a computational tool for sequence-based prediction and characterization of short disorder-to-order transitioning binding regions in proteins. Bioinformatics 2012; 28:i75-i83; PMID:22689782; http://dx.doi.org/10.1093/bioinformatics/bts209
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 2009; 37:W202-8; PMID:19458158; http://dx.doi.org/10.1093/nar/gkp335
