Abstract
The transcriptomics approach to study gene expression in root hairs from M. truncatula has shed light on the developmental events during rhizobial infection and the underlying hormone responses. This approach revealed the induction of several cyclins and an aurora kinase which suggests that the cell-division machinery plays a role in rhizobial infection. Changes in the cell cycle in plants are governed by hormones, in particular auxin and cytokinin. Through gene expression and genetic analyses, we have shown auxin plays a role during rhizobial infection. Here we provide further analysis of the data showing the induction of a set of cytokinin signaling components. These include genes encoding 2 cytokinin-activating enzymes, the cytokinin receptor CRE1, and 5 type-A cytokinin response regulators. We discuss the possible interactions between auxin and cytokinin signaling during the infection process. We also consider a potential role for cytokinin signaling in rhizobial attachment.
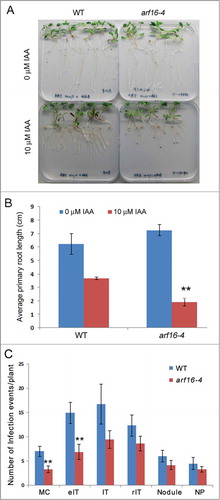
Recently, Breakspear et al.Citation1 characterized gene expression responses prior to and during rhizobial infection and in response to Nod factors in root hairs of Medicago truncatula seedlings. This single-cell type approach offered increased specificity and dramatically enhanced sensitivity of gene detection, the latter being further enhanced by the use of the hyperinfected mutant sickle (skl). It thereby allowed detection of genes that have not otherwise been detected in gene expression studies of nodulation and also allowed the detection of more subtle transcriptional changes that would have otherwise been missed in studies using whole nodulated roots. This was particularly useful in detecting changes in cell-cycle related genes, including the induction of several cyclins, an aurora kinase, and genes involved in hormone biosynthesis and signaling. In particular it led to the discovery of an auxin response factor (ARF), ARF16a, which was specifically induced in root hairs undergoing infection. arf16a mutants showed a reduced number of rhizobial infections revealing the need for regulation of auxin responses in this process. Three alleles were reported in the original study, here we report a fourth allele, arf16a-4, and show that it has a similar phenotype as the other alleles: hypersensitivity to auxin in a root growth assay (), and reduced number of microcolonies and elongating infection threads ().
Figure 1. Auxin response and nodulation phenotype of M. truncatula arf16a-4 mutant. (A) and (B) The inhibition of primary root growth by 10µM indole acetic acid in the wild type (R108) and arf16a-4 (NF4811). The picture (A) and histograms (B) show plants 14 d after germination. (C) Quantification of different stages of infection and development of nodule primordia in the wild type and arf16a-4 mutants 7 dpi with S. meliloti. Infection events and nodule primordia were scored 7 dpi with S. meliloti 1021 carrying pXLGD4 (LacZ) after LacZ staining. IT, fully elongated infection thread in root hair; eIT, elongating infection thread in root hair; MC, microcolony; rIT, ramified infection thread in cortex; NP, nodule primordium. Bar = SE. Significant (Student's t-test) differences between the wild type and mutant are marked with asterisks (**P < 0.01).
The role of auxin signaling in rhizobial infection is not known. One potential role for auxin is in the control of the cell-division machinery which has been found to be associated with infection.Citation1 The hormone cytokinin is widely accepted to act in concert with auxin, often acting to counter or antagonize auxin signaling to establish important signaling fields in different developmental contexts.Citation2 To further explore this proposition we have re-analyzed the data set from Breakspear et al.Citation1 to consider cytokinin responses.
Several components of cytokinin signaling were found to respond to rhizobia and/or Nod factors in root hairs (). Five Type-A cytokinin response regulators were found to be increased: MtRRA2, MtRRA3 (previously MtRR8),Citation3 MtRRA4 (previously MtRR4);Citation4 MtRRA8, and MtRRA10 (previously MtRR11).Citation5 The gene names are according to Heyl et al.,Citation6 personal communication M. Brault and F. Frugier. The increased expression seen with MtRRA8 matches well with an early report that showed that the promoter of the Arabidopsis ortholog ARR5 was expressed in infected root hairs of M. truncatula.Citation7 Type-A response regulators, which act as a central part of a 2-component signaling pathway, are induced by cytokininCitation8,9 and have been shown to act as negative regulators of cytokinin signaling.Citation10 Notably, no type-B response regulators, which mediate the downstream effects of auxin signaling, were found to be induced. This is similar to cytokinin treatments, which induce type A response regulators, but not type B (discussed in D'Agostino et al. 2000).Citation9 Along with these, the cytokinin receptor gene CRE1, was also induced (). Mutants for LHK1 (the Lotus japonicus ortholog of CRE1) have a strong delay in the onset of cell divisions, and nodules that do form are misshapenCitation11,12 and CRE1-knockdown roots form fewer nodules.Citation4 Consistent with these results, promoter-GUS analysis of LHK1 in L. japonicus revealed expression in root hairs associated with infection sites.Citation12 We also found that 2 members of the LONELY GUY gene family which encode an enzyme required for cytokinin-activation were also upregulated (), further suggesting that levels of active cytokinin may be increasing during infection. Interestingly, like CRE1, these genes LOG1 and LOG2 are also induced in nodulation and are required for nodule organogenesis.Citation13 Together these data suggest that cytokinin is being activated in root hairs during the onset of infection and is being perceived through CRE1 to regulate cytokinin signaling.
Table 1. Regulation of cytokinin-related genes in isolated root hairs in response to rhizobial inoculation and application of Nod factors (NFs)
Cytokinin as a Counterpoint to Auxin
The interactions between auxin and cytokinin signaling have been relatively well-studied, and have mainly been found to be antagonistic. For instance, studies of the Arabidopsis meristem have shown that domains of auxin and cytokinin signaling are mutually exclusive.Citation14 The large number of RRAs induced and the apparent absence of increased expression of RRBs may indicate that although cytokinin signaling is active, auxin signaling outcomes prevail in cells undergoing infection.
One possible explanation for the activation of cytokinin signaling during infection may be to antagonize auxin signaling. But how might this work in the case of nodulation? The basis of cytokinin-auxin interactions is not completely understood, but ethylene, which is known to crosstalk with both cytokinin and auxin, and is a major regulator of nodulation, may be part of the mechanism. One major outcome of cytokinin signaling is stabilization of the ACS5 enzyme mediating the rate limiting step of ethylene biosynthesis.Citation15 Our data showed that ethylene signaling represses infection-related gene expression,Citation1 which is presumably due to interference with Nod factor signaling.Citation16 Therefore one role of cytokinin signaling may be to generate ethylene to limit infections, consistent with the observation that the lhk1 mutant has greatly increased numbers of infection threads.Citation11 In further agreement with this hypothesis, increased expression of CRE1 and LOG1 in response to rhizobial inoculation was only observed in skl ().
In addition, ethylene may in turn influence auxin. Indeed, low levels of ethylene have been shown to promote auxin biosynthesis through WEAK ETHYLENE INSENSITIVE1 (WEI1) and WEI2.Citation17 However, the Medicago orthologues of WEI1 and WEI2 are not regulated following rhizobial inoculation or Nod factor treatment (not shown), and since the ethylene insensitive mutant skl becomes hyperinfected, it seems unlikely that ethylene induction of auxin biosynthesis is required for infection. Considering this, the interaction between cytokinin and auxin during infection, if it does occur, may be more direct. Cytokinin and auxin signaling mutants are needed to help address this question.
Cytokinin as a Regulator of Bacterial Attachment
Recently cytokinin has been implicated in bacterial attachment. Agrobacterium tumefaciens produces cytokinins, which were shown to activate the Arabidopsis Type-A cytokinin response regulator ARR3Citation18 which was associated with decreased expression of the Myb Family Transcription Factor 1 (MTF1). Mutant mtf1 plants were shown to have increased A. tumefaciens attachment and improved transformation efficiency. The authors further demonstrated that mutating the cytokinin receptors CRE1 and AHK3 increased the expression of MTF1 and reduced the transformation efficiency. Notably we find that the Medicago ortholog of MTF1 is repressed in the hyperinfected skl mutant at the onset of infection (). This presents an unexpected role through which cytokinin may act during the early stages of the symbiosis, and may add another layer of complexity to cytokinin's role in nodulation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Breakspear A, Liu C, Roy S, Stacey N, Rogers C, Trick M, Morieri G, Mysore KS, Wen J, Oldroyd GE, et al. The root hair “infectome” of medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 2014; 26:4680–701; PMID:25527707; http://dx.doi.org/10.1105/tpc.114.133496
- Schaller GE, Bishopp A, Kieber JJ. The yin-yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell 2015; 27:44–63; http://www.plantcell.org/cgi/doi/10.1105/tpc.114.133595; PMID:25604447
- Saur IM, Oakes M, Djordjevic MA, Imin N. Crosstalk between the nodulation signaling pathway and the autoregulation of nodulation in Medicago truncatula. New Phytol 2011 190:865–74; PMID:21507004; http://dx.doi.org/10.1111/j.1469-8137.2011.03738.x
- Gonzalez-Rizzo S, Crespi M, Frugier F. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 2006; 18:2680–93; PMID:17028204; http://dx.doi.org/10.1105/tpc.106.043778
- Op den Camp RH, De Mita S, Lillo A, Cao Q, Limpens E, Bisseling T, Geurts R. A phylogenetic strategy based on a legume-specific whole genome duplication yields symbiotic cytokinin type-A response regulators. Plant Physiol 2011 157:2013–22; PMID:22034625; http://dx.doi.org/10.1104/pp.111.187526
- Heyl A, Brault M, Frugier F, Kuderova A, Lindner AC, Motyka V, Rashotte AM, Schwartzenberg KV, Vankova R, Schaller GE. Nomenclature for members of the two-component signaling pathway of plants. Plant Physiol 2013 161:1063–5; PMID:23324541; http://dx.doi.org/10.1104/pp.112.213207
- Lohar DP, Schaff JE, Laskey JG, Kieber JJ, Bilyeu KD, Bird DM. Cytokinins play opposite roles in lateral root formation, and nematode and Rhizobial symbioses. Plant J 2004; 38:203–14; PMID:15078325; http://dx.doi.org/10.1111/j.1365-313X.2004.02038.x
- Taniguchi M, Kiba T, Sakakibara H, Ueguchi C, Mizuno T, Sugiyama T. Expression of Arabidopsis response regulator homologs is induced by cytokinins and nitrate. FEBS Lett 1998; 429:259–62; PMID:9662428; http://dx.doi.org/10.1016/S0014-5793(98)00611-5
- D'Agostino IB, Deruère J, Kieber JJ. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 2000 124:1706–17; PMID:11115887; http://dx.doi.org/10.1104/pp.124.4.1706
- Lee DJ, Park JY, Ku SJ, Ha YM, Kim S, Kim MD, Oh MH, Kim J. Genome-wide expression profiling of ARABIDOPSIS RESPONSE REGULATOR 7(ARR7) overexpression in cytokinin response. Mol Genet Genomics 2007; 277:115–37; PMID:17061125; http://dx.doi.org/10.1007/s00438-006-0177-x
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K. A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 2007; 315:101–4; PMID:17110535; http://dx.doi.org/10.1126/science.1132514
- Held M, Hou H, Miri M, Huynh C, Ross L, Hossain MS, Sato S, Tabata S, Perry J, Wang TL, et al. Lotus japonicus cytokinin receptors work partially redundantly to mediate nodule formation. Plant Cell 2014; 26:678–94; PMID:24585837; http://dx.doi.org/10.1105/tpc.113.119362
- Mortier V, Wasson A, Jaworek P, De Keyser A, Decroos M, Holsters M, Tarkowski P, Mathesius U, Goormachtig S. Role of LONELY GUY genes in indeterminate nodulation on Medicago truncatula. New Phytol 2014; 202:582–93; PMID:24443934; http://dx.doi.org/10.1111/nph.12681
- Cheng ZJ, Wang L, Sun W, Zhang Y, Zhou C, Su YH, Li W, Sun TT, Zhao XY, Li XG, et al. Pattern of Auxin and Cytokinin Responses for Shoot Meristem Induction Results from the Regulation of Cytokinin Biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol 2013; 161:240–51; PMID:23124326; http://dx.doi.org/10.1104/pp.112.203166
- Vogel JP, Woeste KE, Theologis A, Kieber JJ. Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc Natl Acad Sci U S A 1998; 95:4766–71; PMID:9539813; http://dx.doi.org/10.1073/pnas.95.8.4766
- Oldroyd GE, Engstrom EM, Long SR. Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell 2001; 13:1835–49; PMID:11487696; http://dx.doi.org/10.1105/tpc.13.8.1835
- Ivanchenko MG, Muday GK, Dubrovsky JG. Ethylene-auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana. Plant J 2008; 55:335–47; PMID:18435826; http://dx.doi.org/10.1111/j.1365-313X.2008.03528.x
- Sardesai N, Lee LY, Chen H, Yi H, Olbricht GR, Stirnberg A, Jeffries J, Xiong K, Doerge RW, Gelvin SB. Cytokinins secreted by Agrobacterium promote transformation by repressing a plant myb transcription factor. Sci Signal 2013 6(302):ra100; PMID:24255177
