Abstract
Two independent studies have shown that the cell wall of pollen tubes from tobacco and tomato species contained fucosylated xyloglucan (XyG). These findings are intriguing as many reports have shown that XyG of somatic cells of these species is not fucosylated but instead is arabinosylated. In order to produce fucosylated XyG, plants must express a functional galactoside α-2-fucosyltransferase. Here, using a bioinformatics approach, we show that several candidate genes coding for XyG fucosyltransferases are present in the genome of coffee and several Solanaceae species including tomato, tobacco, potato, eggplant and pepper. BLAST and protein alignments with the 2 well-characterized XyG fucosyltransferases from Arabidopsis thaliana and Pisum sativum revealed that at least 6 proteins from different Solanaceae species and from coffee displayed the 3 conserved motifs required for XyG fucosyltransferase activity.
Abbreviations
| AA | = | amino acid |
| AGP | = | arabinogalactan proteins |
| FUT | = | fucosyltransferase |
| XyG | = | xyloglucan |
Two recent independent studies have shown that the pollen tube cell wall of tobacco (Nicotiana alata and N. tabacum) and wild (Solanum peruvianum and S. pimpinellifolium) and domesticated (S. lycopersicum) tomato species contains fucosylated xyloglucan (XyG).Citation1,2 We have previously reported in Arabidopsis thaliana that the XyG of pollen tubes was also highly fucosylated,Citation3 suggesting an important role of the XyG fucosylation in the pollen tube growth. In all the investigated species, the main fucosylated motif, detected by MALDI-TOF MS after endo-glucanase digestion of the XyG, was the mono-acetylated XXFG Citation1-Citation3 according to the nomenclature proposed by Fry et al.Citation4 Interestingly, in the cell wall of vegetative organs and/or suspension-cultured cells from plants of the Asterid clade (including the Solanaceae),Citation5 XyG lacks fucosyl residues and instead exhibits an arabinoXyG-type, due presumably to an adaptative and/or selective diversification.Citation2,6-10 Thus, pollen tubes and vegetative organs from the Solanaceae have structurally divergent XyG. Despite the fact that RNA-Seq analyses of N. alata pollen tubes could not detect any XyG fucosyltransferases,Citation1 pollen tubes from Solanaceae species must have a specific set of functional XyG fucosyltransferases (FUTs).
In order to find putative XyG FUTs, we used the Basic Local Alignment Search Tool (BLAST) with the sequence of AtFUT1, the well-characterized Arabidopsis XyG galactoside α-2-fucosyltransferase,Citation11 as the reference. AtFUT1 was characterized as a galactoside α-2-fucosyltransferase able to transfer L-fucosyl residues to galactosylated XyG.Citation11,12 Two BLAST algorithms were used: the BLAST software of the Sol Genomic Network (http://solgenomics.net/tools/blast/) and the BLAST tool of the Uniprot database (http://www.uniprot.org/blast/). The BLAST performed in the Sol genomic Network used the sequence of AtFUT1 against the following databases: the Tomato proteins ITAG (release 2.3), the potato PGSC DM v3.4 protein sequences, the Nicotiana benthamiana genome v0.4.4 predicted proteins, the Capsicum annuum cv CM334 genome protein sequences, the Eggplant (Solanum melongena) Genome protein sequences (release 2.5.1), the Nicotiana tabacum TN90 protein sequences and the Coffea canephora protein sequences v1.0.
By searching amino acid (AA) sequence similarities with AtFUT1Citation11 the BLAST retrieved several proteins including PsFT1, another characterized XyG FUT from Pisum sativum.Citation13 PsFT1 showed 59.2% of AA identity with AtFUT1 (). When the BLAST was performed against the database "tomato Proteins ITAG release 2.3," 3 predicted proteins from S. lycopersicum coded by the Solyc07g047920.1, Solyc06g061210.2 and Solyc03g115830.1 genes were tagged as putative fucosyltransferasesCitation2 (). These 3 predicted proteins from S. lycopersicum displayed 57.5%, 55.3% and 53.9% of AA identity with AtFUT1, respectively. Two putative FUTs were also found in the potato protein sequences, corresponding to the genes PGSC0003DMG400033900 and PGSC0003DMG400003781. These proteins showed 55.8% and 59.2% of AA identity with the sequence of AtFUT1 (). Two other putative FUTs were found in the genome of Solanum melongena with 57.4 and 57.9% of AA identity with AtFUT1. The search in the database of predicted proteins of N. benthamiana retrieved 2 sequences displaying 56.5 and 55.6% of AA identity with AtFUT1 (). In the Nicotiana tabacum genome, 4 genes are predicted to encode FUTs. The proteins showed about 56% of AA identity with the sequence of AtFUT1 (). In the Capsicum annuum genome, 2 putative FUTs coded by Ca03g29900 and Ca06g13300 were also found. These putative XyG FUTs from pepper showed 56.7 and 54.5% of AA identity with AtFUT1 (). Finally, 4 putative XyG FUTs were found in Coffea canephora showing between 53.7 and 58.6 % of AA identity with AtFUT1 ().
Table 1. Comparison of the length, identity and similarity of amino acid sequences among 2 galactoside 2-α-L-fucosyltransferases from Arabidopsis thaliana (AtFUT1) and Pisum sativum (PsFT1) and 15 putative galactoside 2-α-L-fucosyltransferases from Solanum lycopersicum, Solanum tuberosum, Solanum melongena, Nicotiana benthamiana, Nicotiana tabacum and Capsicum annuum. The sequences from all the Solanaceae species were retrieved as a result of sequence similarity search (BLAST) against AtFUT1, using the blast software of the Sol Genomic Network.Citation14,15 Identity and similarity were calculated using the BLOSUM 62 matrix. Names, Sequence ID, function or annotation have been retrieved from UniProt or from the Sol Genomic Network.
AtFUT1 and PsFT1 exhibit a transmembrane domain that is also present in the 15 putative XyG FUTs retrieved in S. lycopersicum, S. tuberosum N. benthamiana, N. tabacum, C. annuum and C. canephora as determined by TMHMMCitation16 (). This is consistent with FUTs being type-II membrane-bound proteins.Citation10,11 However, no transmembrane domain was found neither in the putative XyG FUT from S. melongena nor in 2 of the putative XyG FUTs from C. canephora (CcFUTc and CcFUTd) ().
Figure 1. Sequences of the type-II transmembrane domain and the 3 peptide motifs shared by the 2 characterized α-2-fucosyltransferases, AtFUT1 and PsFT1, and the putative α-2-fucosyltransferases from tomato (Sl), potato (St), eggplant (Sm), 2 species of tobacco (Nb and Nt), pepper (Ca) and coffee (Cc). Conserved residues are represented by white letters in black background; highly similar AA are shaded in gray. Numbers inside angle brackets indicate the number of AA between 2 motifs. Proteins are named according to the code used in .
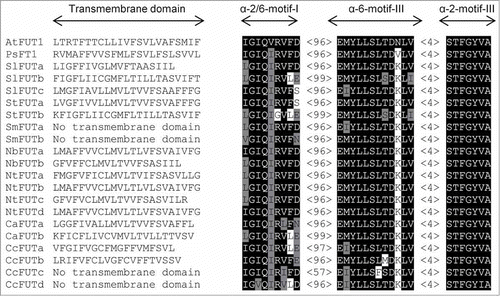
It has been shown that all known α-1,2- and α-1,6-FUTs contain 3 conserved motifs, the α-2/6-motif I, the α-2-motif III and the α-6-motif III.Citation17,18 Looking in detail at the peptide motifs shared by these putative XyG FUTs (), we can notice that the α-2-motif III is strictly conserved in the 19 predicted proteins and that the α-6-motif III and the α-2-motif III are separated by 4 AA in all the sequences. The α-6-motif III from AtFUT1 and PsFT1 is also conserved with the exception that the asparagine (N) is replaced by a valine (V) in pea (). In the other putative XyG FUTs from tomato, potato, eggplant, tobacco, pepper and coffee, the α-6-motif III is also very similar to the one found in AtFUT1. Particularly, SlFUTa from tomato, StFUTa from potato, 6 proteins from tobacco, 2 from pepper and SmFUTb from eggplant contained a motif composed of 12 AA. Among them, 11 are identical when compared to the α-6-motif III from Arabidopsis or pea. Only 1 AA differed: the asparagine (N, in Arabidopsis) or the valine (V, in pea) is replaced by a lysine (K) in the Solanaceae sequences ().
The sequences of the α-2/6-motif I from AtFUT1 and PsFT1 is also strictly conserved with the exception that the hydrophobic V in Arabidopsis that is replaced by another hydrophobic AA, isoleucine (I) in pea and the other sequences (). The motif from pea showed 100% of AA identity with 2 proteins from tobacco (NbFUTa and NtFUTd), 100% of similarity with 1 from tomato (SlFUTa), 4 from tobacco (NbFUTb, NTFUTa, NtFUTb and NtFUTc) and 1 from pepper (CaFUTa) (). In these proteins, with the exception of pepper, the first AA of the motif (I in pea) is replaced by a leucine (L). One sequence from potato (StFUTa) and 1 from tomato (SlFUTc) also contained the motif with a high degree of similarity with PsFT1 with the exception that the last AA of the motif, aspartic acid (D), is replaced by a serine (S) (). These 9 sequences from potato, tobacco, tomato and pepper have also the same gap (96 AA) between the α-2/6-motif I and the α-6-motif III found in AtFUT1 and PsFT1 (). In contrast, the 5 sequences showing the less AA similarity with AtFUT1 and PsFT1 (i.e., SlFUTb, StFUTb, CaFUTb, CcFUTa and CcFUTc) did not contain the 96 AA gap between the α-2/6-motif I and the α-6-motif III () but displayed a larger gap (97 or 99 AA) or a smaller one of 57 AA ().
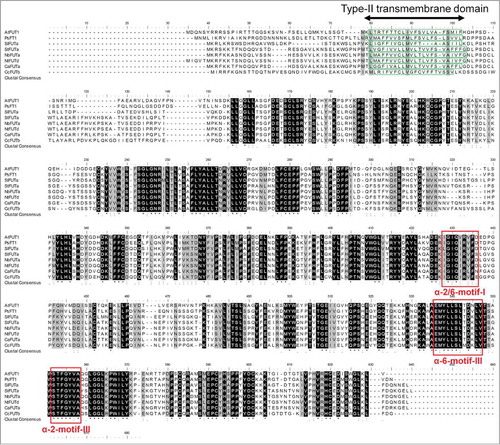
In order to visualize the homology of these putative XyG FUTs, we aligned AtFUT1, PsFT1 and the 6 best predicted XyG FUTs from S. lycopersicum, S. tuberosum, N. benthamiana, N. tabacum and C. annuum and C. canephora with the ClustalW algorithm (). The proteins were chosen based on the similarities highlighted in (presence of the transmembrane domain, identity of the XyG FUT motifs and the gap between the motifs). The alignment revealed that these proteins shared strong similarities () all along their sequences suggesting that they probably belong to the XyG FUT family.
Figure 2. Amino acid (AA) sequence alignments using the ClustalW algorithm. Black and gray shadings represent conserved and highly similar AA, respectively, among the best candidate for putative fucosyltransferases from S. lycopersicum (SlFUTa), S. tuberosum (StFUTa), N. benthamiana (NbFUTa), N. tabacum (NtFUTd), C. annuum (CaFUTa), C. canephora (CcFUTb) and the 2 characterized galactoside 2-α-L-fucosyltransferases from Arabidopsis thaliana and Pisum sativum: AtFUT1 and PsFT1. The type-II transmembrane domains are boxed in green dashed lines. The conserved motifs (α-2/6-motif I, α-6-motif III and α-2-motif III) found in all known α-1,2-fucosyltransferases are boxed in red solid lines. Proteins are named according to the code presented in .
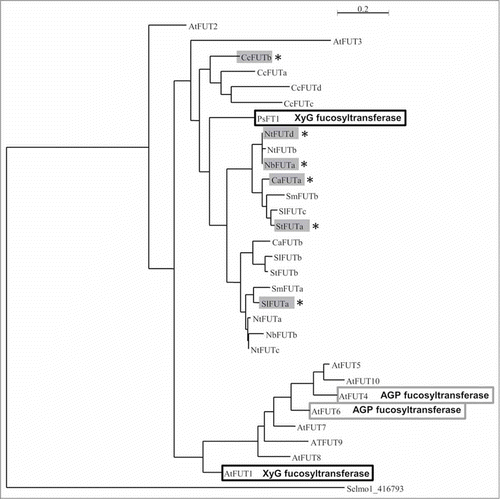
XyG is not the only polymer containing fucosyl residue linked to galactose in plant cell walls. Among the 10 members of the FUT family in Arabidopsis, 2 of them AtFUT4 and AtFUT6 were characterized as arabinogalactan-protein (AGP) FUT.Citation19 In order to discriminate if the selected proteins presented in may act preferentially on AGPs or on XyG, a phylogenic analysis was performed. To do so, a predicted XyG FUT from Selaginella moellendorffii was used as an outgroup sequence (gene name: GT37A1–1, UniProt D8S0F5). All the sequences from the and the 10 sequences of the Arabidopsis FUT family were analyzed using the methods described by Dereeper et al. (2008) (phylogeny.fr).Citation20 Sequences were aligned with MUSCLE (full processing mode), the curation was performed using the Gblocks program to eliminate poorly aligned positions and divergent regions. The phylogeny was performed with PhyML (model: WAG; Statistical: alrt; Number of categories: 4; Gamma: estimated; invariable sites: estimated). Finally the tree rendering was run with Tree dyn and is presented in . Firstly, it appears that the sequences are classified based on the species they belong to except for AtFUT2 and AtFUT3. From the bottom to the top: Arabidopsis, Solanaceae, pea and the coffee FUT families. It appears also that the AtFUT family can be subdivided into AtFUT1, AtFUT2 and AtFUT3, each in their own group, and AtFUT4 to 10 in a larger group more related to AGP FUTs (). Interestingly, 5 out of the 6 proteins (NtFUTd, NbFUTa, CaFUTa, StFUTa, SlFUTa) aligned with AtFUT1 and PsFT1 () belong to a large subgroup including PsFT1. The last sequence, from coffee (CcFUTb) belongs to another sub group ().
Figure 3. Phylogenic tree showing the relationships between the AtFUT family and the 19 sequences retrieved in the coffee and the Solanaceae genomes. The tree was generated with MUSCLE for alignment (full processing mode), Gblocks for curation and then built with PhyML (model: WAG; Statistical: alrt; Number of categories: 4; Gamma: estimated; invariable sites: estimated) and rendered with Tree dyn. Black boxed sequences correspond to XyG fucosyltransferases, empty gray boxes correspond to AGP fucosyltransferases and painted gray boxes with * correspond to the sequences used for the alignment presented in . A. thaliana (At), S. lycopersicum (Sl), S. tuberosum (St), N. benthamiana (Nb), N. tabacum (Nt), C. annuum (Ca), C. canephora (Cc), P. sativum (Ps), Selaginella moellendorffii (Selmo).
The recent discovery of fucosylated XyG in tomato and tobacco pollen tubes implied the presence of functional XyG FUTs. The search of proteins closely related to the well-characterized galactoside α-2-fucosyltransferase, AtFUT1,Citation11,21 retrieved, in addition to PsFT1,Citation13 19 predicted proteins in S. lycopersicum, S. melongena, S. tuberosum, N. benthamiana, N. tabacum, C. annuum, and C. canephora with the 3 motifs found in all known α-1,2- and α-1,6-fucosyltransferases.Citation17,18 Six among these 19 putative XyG FUTs exhibited very strong similarities with AtFUT1 and/or PsFT1 and they are likely to be good candidates to perform the transfer of a fucosyl residue on the galactose of XyG. However, as bioinformatics cannot predict with absolute certainty the donor and the acceptor substrate for these enzymes, experimental evidence is needed to verify in more detail the expression pattern of these genes in pollen grains and pollen tubes and to assess the biochemical functions of these proteins.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the “Haute-Normandie” region, the Research Network 'Végétal, Agronomie, Sol, Innovation' (VASI) and the University of Rouen.
References
- Lampugnani ER, Moller IE, Cassin A, Jones DF, Koh PL, Ratnayake S, Beahan CT, Wilson SM, Bacic A, Newbigin E. In vitro grown pollen tubes of Nicotiana alata actively synthesise a fucosylated xyloglucan. PLoS ONE 2013; 8:e77140; PMID:2411621; http://dx.doi.org/ 10.1371/journal.pone.0077140
- Dardelle F, Le Mauff F, Lehner A, Loutelier-Bourhis C, Bardor M, Rihouey C, Causse M, Lerouge P, Driouich A, Mollet JC. Pollen tube cell walls of wild and domesticated tomatoes contain arabinosylated and fucosylated xyloglucan. Ann Bot 2015; 115:55- 66; PMID:25434027; http://dx.doi.org/10.1093/aob/mcu218
- Dardelle F, Lehner A, Ramdani Y, Bardor M, Lerouge P, Driouich A, Mollet JC. Biochemical and immunocytological characterizations of Arabidopsis thaliana pollen tube cell wall. Plant Physiol 2010; 153:1563-76; PMID:20547702; http://dx.doi.org/10.1104/pp.110.158881
- Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau JP, Kato Y, Pérez Lorences E, Maclachlan GA, McNeil M, et al. An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol Plant 1993; 89:1-3. http://dx.doi.org/10.1111/j.1399-3054.1993.tb01778.x
- The Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc 2009; 161:105-21. http://dx.doi.org/10.1111/j.1095-8339.2009.00996.x
- York WS, Kumar Kolli VS, Orlando R, Albersheim P, Darvill AG. The structures of arabinoxyloglucans produced by solanaceous plants. Carbohydr Res 1996; 285:99-128; PMID:9011379
- Hoffman M, Jia Z, Pena MJ, Cash M, Harper A, Blackburn AR, Darvill A, York WS. Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr Res 2005; 340:1826-40; PMID:15975566; http://dx.doi.org/ 10.1016/j.carres.2005.04.016
- Jia Z, Cash M, Darvill AG, York WS. NMR characterization of endogenously O-acetylated oligosaccharides isolated from tomato (Lycopersicon esculentum) xyloglucan. Carbohydr Res 2005; 340:1818-25; PMID:15927168; http://dx.doi.org/10.1016/j.carres.2005.04.015
- Schultink A, Liu L, Zhu L, Pauly M. Structural diversity and function of xyloglucan sidechain substituents. Plants 2014; 3:526-42. http://dx.doi.org/10.3390/plants3040526
- Popper ZA, Tuohy MG. Beyond the green: understanding the evolutionary puzzle of plant and algal cell walls. Plant Physiol 2010; 153:373-83; PMID:20421458; http://dx.doi.org/ 10.1104/pp.110.158055
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K. Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 1999; 284:1976-9; PMID:10373113; http://dx.doi.org/ 10.1126/science.284.5422.1976
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD. The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc Natl Acad Sci USA 2002; 99:3340-5; PMID:11854459; http://dx.doi.org/10.1073/pnas.052450699
- Faik A, Bar-Peled M, DeRocher AE, Zeng W, Perrin RM, Wilkerson C, Raikhel NV, Keegstra K. Biochemical characterization and molecular cloning of an α-1,2-fucosyltransferase that catalyse the last step of cell wall xyloglucan biosynthesis in pea. J Biol Chem 2000; 275:15082-9; PMID:10747946; http://dx.doi.org/ 10.1074/jbc.M000677200
- Mueller LA, Solow TH, Taylor N, Skwarecki B, Buels R, Binns J, Lin C, Wright MH, Ahrens R, Wang Y, et al. The SOL Genomics Network: a comparative resource for Solanaceae biology and beyond. Plant Physiol 2005; 138:1310-7; PMID:16010005; http://dx.doi.org/10.1104/pp.105.060707
- Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S, Fischer-York T, Pujar A, Leto J, Gosselin J, Mueller LA. The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res 2011; 39:D1149-55; PMID:20935049; http://dx.doi.org/ 10.1093/nar/gkq866
- Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001; 305:567-80; PMID:11152613; http://dx.doi.org/10.1006/jmbi.2000.4315
- Breton C, Oriol R, Imberty A. Conserved structural features in eukaryotic and prokaryotic fucosyltransferases. Glycobiology 1998; 8: 87-94; PMID:9451017
- Oriol R, Mollicone R, Cailleau A, Balanzino L, Breton C. Divergent evolution of fucosyltransferase genes from vertebrates, invertebrates and bacteria. Glycobiology 1999; 9:323-34; PMID:10089206
- Wu Y, Williams M, Bernard S, Driouich A, Showalter AM, Faik A. Functional identification of two nonredundant Arabidopsis α (1,2) fucosyltransferases specific to arabinogalactan proteins. J Biol Chem 2010; 285: 13638-45; PMID:24667545; http://dx.doi.org/10.1074/jbc.M110.102715
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008; 36: W465-9; PMID:18424797; http://dx.doi.org/10.1093/nar/gkn180
- Perrin RM, Jia Z, Wagner TA, O'Neill MA, Sarria R, York WS, Raikhel NV, Keegstra K. Analysis of xyloglucan fucosylation in Arabidopsis. Plant Physiol 2003; 132:768-78; PMID:12805606; http://dx.doi.org/10.1104/pp.102.016642
