Abstract
Splicing and alternative splicing (AS) are widespread co- and post-transcriptional regulatory processes in plants. Recently, we characterized genome-wide AS landscapes and virus-induced AS patterns in Brachypodium distachyon (Brachypodium), a C3 model grass. Brachypodium plants infected with Panicum mosaic virus (PMV) alone or in mixed infections with its satellite virus (SPMV) were used for high-throughput, paired-end RNA sequencing. Here, using gene attributes of ∼5,655 intronless genes, ∼13,302 constitutively spliced, and ∼7,564 alternatively spliced genes, we analyzed the influence of genomic features on splicing incidence and AS frequency. In Brachypodium, gene length, coding sequence length, and exon and intron number were positively correlated to splicing incidence and AS frequency. In contrast, exon length and the percentage composition of GC (%GC) content were inversely correlated with splicing incidence and AS frequency. Although gene expression status had little correlation with splicing occurrence per se, it negatively correlated to AS frequency: i.e., genes with ≥5 alternatively spliced transcripts were significantly less expressed compared to genes encoding <5 alternative transcripts. Further gene set enrichment analysis uncovered unique functional relationships among nonspliced, constitutively spliced and alternatively spliced genes.
Introduction
About 95% of genes in the human genome are alternatively spliced, and splicing-related processes underlie upwards of 15% of human genetic diseases.Citation1,2 Recent studies using next-generation RNA sequencing demonstrated that alternative splicing (AS) is also prevalent among plants with about 60%, 50% and 35% of the expressed genes in Arabidopsis, soybean and rice being alternatively spliced.Citation3-6 Several plant genes that function in flowering processes, circadian clock and defense are also known to be alternatively spliced.Citation7,8 Furthermore, AS processes are developmentally regulated and modulated during stresses.Citation7,8 Alternative splicing is thought to primarily influence transcriptome and proteome diversity and to provide plasticity in adapting to stress. However, the extent and influence of genomic attributes on AS processes, and the functional relevance of the thousands of AS transcript variants is yet to be investigated.
Brachypodium distachyon (Brachypodium) is a temperate C3 grass that is being used as a model plant to investigate several aspects of grass biology including cell-wall composition, growth and development, and biotic and abiotic stress responses.Citation9,10 Brachypodium is a diploid plant with a small genome (272 Mbp) that is highly collinear to the genomes of key agronomic grasses including wheat, barley, rice and maize.Citation11 We have established Brachypodium and Panicum mosaic virus (PMV) and its satellite virus (SPMV)Citation12-14 as a pathosystem to characterize genome-wide AS landscapes and the occurrence of differential splicing during virus infection.Citation15 Approximately 42% of Brachypodium multi-exonic genes are alternatively spliced. Among the AS events, intron retention events predominated and exon skipping AS events were the least frequent. Here, we further analyzed the genomic factors that influence or constrain Brachypodium splicing incidence and AS frequency, and determined unique functional traits of the different splicing groups and non-spliced genes.
Genomic Attributes and Correlation with Splicing Incidence and Frequency
It is thought that pre-mRNA splicing and recognition of splice sites in plants follows the intron definition model, as opposed to the exon definition model in animals.Citation16 However, the influence of genomic features on splicing incidence and frequency in plants remains largely unknown. To analyze such relationships in the Brachypodium spliceome, we performed statistical analysis for correlations among 8 Brachypodium genomic attributes including gene length, coding sequence (CDS) length, exon number, exon length, intron number, intron length, the %GC content and expression of 5,655 intron less genes, 13,302 constitutively spliced (non-AS), and 7,564 alternatively spliced genes. The expression of the different genes was determined from our RNA-seq analysis.Citation15 The frequency of AS in Brachypodium genes ranged from 2 to 11 events. To detect constraints affecting splicing frequency among genes, we divided the 7,564 AS genes into low (<5 isoforms) and high (≥5 isoforms) frequency. By this criterion, about 6,818 genes were low AS frequency, while 746 genes were high AS frequency. Statistical analyses were performed using the SAS Generalized Linear Method and applying a Tukey's Studentized Range (HSD) test that corrects for Type I experiment-wise false discovery errors. The square of the Pearson product moment correlation coefficient (R2) was used to determine robust correlations.
We found that gene length, CDS length, exon number and intron number were all strongly and positively correlated (R2 ≥ 0.89, α < 0.01) to splicing incidence among intronless, constitutive and alternatively spliced genes (). Further, splicing frequency within the low and high AS categories was also positively correlated to gene length, CDS length, exon number and intron number (). By contrast, both splicing incidence and frequency were negatively correlated (R2 ≥ 0.64, α < 0.01) to exon length and %GC content (), suggesting that exonic splicing signals and nucleotide composition are critical regulators of pre-mRNA splicing in plants.Citation16 We found little correlation between intron length and gene expression with splicing occurrence among the intronless, constitutively spliced and AS genes (). However, splicing frequency was inversely proportional to gene expression levels (). For example, genes with a greater number of isoforms (≥5, high AS genes) had significantly less expression (α < 0.05) compared to genes with <5 isoforms (low AS genes) (). Thus co-transcriptional processes likely guide AS frequency or vice versa. Finally, we analyzed chromosome-level distribution of the AS events, and their relationship with genome size and number of genes in Brachypodium, rice (Oryza sativa var. japonica), and Arabidopsis. In all 3 plants, we found a strong positive correlation of AS event incidence to number of genes per chromosome () with no chromosome-specific biases, attesting to the co- and post-transcriptional nature of the AS phenomenon. Together, these analyses reveal genomic features that influence splicing processes in plants.
Figure 1. Genomic attributes of intronless, constitutively- and alternatively-spliced genes and relationship with splicing incidence and frequency. (A) Gene length [bp] (B) CDS length [bp] (C) exon number, (D) exon length [bp], (E) intron number, (F) intron length [bp] (G) %GC content, and (H) expression [Fragments per kilobase of transcript per million mapped fragments, FPKM] statistics of intronless genes, constitutively spliced (Non-AS) and alternatively spliced (AS) genes are graphed. Genes with <5 and ≥5 isoforms are designated as low and high AS genes respectively. Pearson correlation coefficient values (R2) among the different groups are indicated. Error bars represent standard error. Statistically significant differences (α < 0.01) are determined by Tukey's Studentized Range test. AS event frequency and correlation to genome size and number of gene models per chromosome in (I) Brachypodium, (J) rice, and (K) Arabidopsis are shown. The genomic features of Brachypodium, rice and Arabidopsis were obtained from Phytozome, IGRSP, and TAIR databases, respectively.
![Figure 1. Genomic attributes of intronless, constitutively- and alternatively-spliced genes and relationship with splicing incidence and frequency. (A) Gene length [bp] (B) CDS length [bp] (C) exon number, (D) exon length [bp], (E) intron number, (F) intron length [bp] (G) %GC content, and (H) expression [Fragments per kilobase of transcript per million mapped fragments, FPKM] statistics of intronless genes, constitutively spliced (Non-AS) and alternatively spliced (AS) genes are graphed. Genes with <5 and ≥5 isoforms are designated as low and high AS genes respectively. Pearson correlation coefficient values (R2) among the different groups are indicated. Error bars represent standard error. Statistically significant differences (α < 0.01) are determined by Tukey's Studentized Range test. AS event frequency and correlation to genome size and number of gene models per chromosome in (I) Brachypodium, (J) rice, and (K) Arabidopsis are shown. The genomic features of Brachypodium, rice and Arabidopsis were obtained from Phytozome, IGRSP, and TAIR databases, respectively.](/cms/asset/7b9f85a5-4797-4b55-9270-4be346831246/kpsb_a_1042640_f0001_oc.gif)
Functional Enrichment and Relationships among Brachypodium Splicing Groups
To define genome-wide functional relationships of genes in the different splicing groups, we took a statistical approach and analyzed gene ontology (GO) functional annotations. Gene set enrichment analysis (SEA) of the different GO terms in biological processes, molecular functions and cellular components was performed for the intronless, non-AS and AS genes using the AgriGO program.Citation17 Gene SEA was performed against a pre-calculated Brachypodium reference genome using χ2 test for significance, followed by false discovery rate (FDR) correction using the Hochberg method (FDR <0.05). We identified about 124 GO terms that were significantly enriched in the different splicing groups. About 76 terms were enriched among all 3 splicing groups (), including biological processes (primary metabolism, transport, gene expression, protein modification) and cellular components (nucleus, cytoplasm and other organelles) (). By narrowing the functional terms that were uniquely enriched among the splicing groups, we found 14, 10 and 7 terms that were unique among intronless, non-AS and AS genes, respectively. These 31 terms were subjected to REVIGO redundancy filtering, and significantly enriched terms were plotted in 2-dimensional, semantic-space scatter plots to visualize relationships among the different terms.Citation18
Figure 2. Gene ontology (GO) analyses of Brachypodium splicing groups. (A) Distribution of significantly enriched (FDR<0.05) GO terms among intronless, non-AS and AS genes in Brachypodium. REVIGO scatter plot visualization of GO terms in (B) biological process. (C) cellular component, and (D) molecular function common among intronless, non-AS and AS gene groups. In the REVIGO plots, bubble color and distance represent uniqueness among the GO terms, where related terms are clustered together with similar color shades. Bubble size represents the relative frequency of the GO term in the underlying GO annotation database, with smaller bubble sizes representing more specific terms.
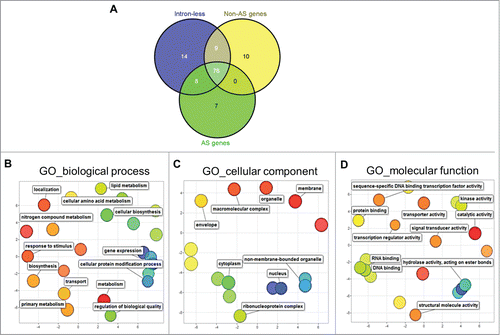
The REVIGO analysis identified biological processes such as photosynthesis (GO:0015979), reproduction (GO:0000003), pollen-pistil interactions (GO:0009875), cell communication (GO:0007154), embryonic development (GO:0009790), and a cellular component thylakoid (GO:0009579) as uniquely enriched among the intronless gene group (). Incidentally, the unique enrichment of photosynthesis- and thylakoid-related functions in the plant intronless genes is consistent with, and further lends support to the modern synthesis of the theory of endosymbiosisCitation19 explaining the origin of plastids from a cyanobacteriumCitation20— a prokaryotic organism whose genes are devoid of introns. Among the non-AS or constitutively-spliced gene groups, biological processes such as response to biotic stimulus (GO:0009628) and abiotic stimulus (GO:0009607), cellular components such as extracellular regions (GO:0005576), cell wall (GO:0005618), plasma membrane (GO:0005886), endoplasmic reticulum (GO:0005783), and intracellular organelle lumen (GO:0070013) were uniquely enriched (). The enrichment of membrane-bound and cell-surface localization terms in the non-AS genes correlates well with signal transduction activity (e. g., receptor-like kinases) required to mediate responses to biotic stimuli.
Figure 3. Unique functions enriched among Brachypodium intronless, constitutively- and alternatively-spliced (AS) genes. Significantly enriched (FDR<0.05) and unique gene ontology (GO) terms in biological process (A–C), and cellular component (D–F) of intronless genes (A and D), non-AS genes (B and F), and AS genes (C and F) are depicted as colored bubbles in REVIGO two-dimensional semantic-space scatter plots. Bubble color and distance represent uniqueness among the GO terms, where related terms are clustered together with similar color shades. Bubble size represents the relative frequency of the GO term in the underlying GO annotation database, with smaller bubble sizes representing more specific terms.
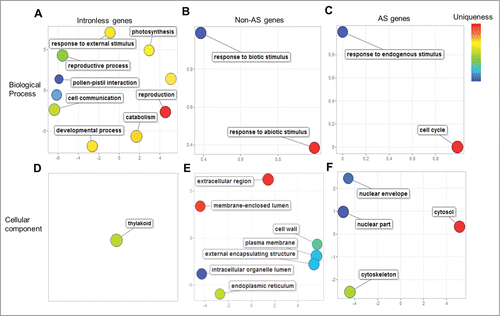
Among the AS genes, unique functions such as response to endogenous stimuli (GO:0009719), cell cycle (GO:0007049), and cellular components such as cytosol (GO:0005829), nuclear part (GO:0044428), nuclear envelope (GO:0005635) and cytoskeleton (GO:0005856) were enriched (). Our analysis suggests that AS genes might respond to endogenous stimuli and cell-cycles in addition to their known responses to biotic and abiotic stresses.
Conclusion
Gene-level architectural differences of the Brachypodium nonspliced, constitutively- and alternatively-spliced genes, and genomic attributes that positively and negatively influenced splicing occurrence and frequency were analyzed. The negative correlation we found between exon length and splicing occurrence suggests that exon splicing signals are also important regulators of pre-mRNA splicing in plants. Although splicing processes and AS genes are implicated in both biotic and abiotic stress responses, our results show that non-AS genes are uniquely enriched in functions related to response to biotic and abiotic stress. In contrast, AS genes are primarily enriched in functions of response to endogenous stimuli and cell cycle. Together, these findings provide novel insight into AS regulation and its functional relevance to plant cellular processes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We appreciate the critical reading and comments of Sonia Irigoyen (Texas A&M AgriLife) and Jesse D. Pyle (Texas A&M) during the preparation of this manuscript.
Funding
This study was partially supported by funds from Texas A&M AgriLife Research Start-up Fund to K.K.M.
References
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature 2008; 456:470-6; PMID:18978772; http://dx.doi.org/10.1038/nature07509
- Krawczak M, Thomas NST, Hundrieser B, Mort M, Wittig M, Hampe J, Cooper DN. Single base-pair substitutions in exon–intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum Mutat 2007; 28:150-8; PMID:17001642; http://dx.doi.org/10.1002/humu.20400
- Marquez Y, Brown JWS, Simpson C, Barta A, Kalyna M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res 2012; 22:1184-95; PMID:22391557; http://dx.doi.org/10.1101/gr.134106.111
- Zhang G, Guo G, Hu X, Zhang Y, Li Q, Li R, Zhuang R, Lu Z, He Z, Fang X, et al. Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res 2010; 20:646-54; PMID:20305017; http://dx.doi.org/10.1101/gr.100677.109
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 2010; 20:45-58; PMID:19858364; http://dx.doi.org/10.1101/gr.093302.109
- Shen Y, Zhou Z, Wang Z, Li W, Fang C, Wu M, Ma Y, Liu T, Kong LA, Peng DL, et al. Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 2014; 26:996-1008; PMID:24681622; http://dx.doi.org/10.1105/tpc.114.122739
- Reddy ASN, Marquez Y, Kalyna M, Barta A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013; 25:3657-83; PMID:24179125; http://dx.doi.org/10.1105/tpc.113.117523
- Staiger D, Brown JWS. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013; 25:3640-56; PMID:24179132; http://dx.doi.org/10.1105/tpc.113.113803
- Brkljacic J, Grotewold E, Scholl R, Mockler T, Garvin DF, Vain P, Brutnell T, Sibout R, Bevan M, Budak H, et al. Brachypodium as a model for the grasses: Today and the future. Plant Physiol 2011; 157:3-13; PMID:21771916; http://dx.doi.org/10.1104/pp.111.179531
- Catalán P, Chalhoub B, Chochois V, Garvin DF, Hasterok R, Manzaneda AJ, Mur LA, Pecchioni N, Rasmussen SK, Vogel JP, et al. Update on the genomics and basic biology of Brachypodium. International Brachypodium Initiative (IBI). Trends Plant Sci 2014; 19:414-8; http://dx.doi.org/10.1016/j.tplants.2014.05.002
- The International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010; 463:763-8; PMID:20148030; http://dx.doi.org/10.1038/nature08747.
- Mandadi KK, Scholthof K-BG. Characterization of a viral synergism in the monocot Brachypodium distachyon reveals distinctly altered host molecular processes associated with disease. Plant Physiol 2012; 160:1432-52; PMID:22961132; http://dx.doi.org/10.1104/pp.112.204362
- Mandadi KK, Pyle JD, Scholthof K-BG. Comparative analysis of antiviral responses in Brachypodium distachyon and Setaria viridis reveal conserved and unique outcomes among C3 and C4 plant defenses. Mol Plant Microbe Interact 2014; 27:1277-90; PMID:25296115; http://dx.doi.org/10.1094/MPMI-05-14-0152-R
- Mandadi KK, Scholthof K-BG. Plant immune responses against viruses: How does a virus cause disease? Plant Cell 2013; 25:1489-505; PMID:23709626; http://dx.doi.org/10.1105/tpc.113.111658
- Mandadi KK, Scholthof K-BG. Genome-wide analysis of alternative splicing landscapes modulated during plant-virus interactions in Brachypodium distachyon. Plant Cell 2015; 27:71-85; PMID:25634987; http://dx.doi.org/10.1105/tpc.114.133991
- Reddy ASN. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol 2007; 58:267-94; PMID:17222076; http://dx.doi.org/10.1146/annurev.arplant.58.032806.103754
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 2010; 38:W64-W70; PMID:20435677; http://dx.doi.org/10.1093/nar/gkq310
- Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLOS ONE 2011; 6:e21800; PMID:21789182; http://dx.doi.org/10.1371/journal.pone.0021800
- Sagan L. On the origin of mitosing cells. J Theor Biol 1967; 14:225-74; http://dx.doi.org/10.1016/0022-5193(67)90079-3
- Baum D. The origin of primary plastids: A pas de deux or a ménage à trois? Plant Cell 2013; 25:4-6; PMID:23371952; http://dx.doi.org/10.1105/tpc.113.109496
