Abstract
In eukaryotes alternative splicing (AS) influences transcriptome and proteome diversity. The mechanism and the genetic components mediating AS during plant-virus interactions are not known. Using RNA sequencing approaches, we recently analyzed the global AS changes occurring in Brachypodium distachyon (Brachypodium) during infections of Panicum mosaic virus (PMV) and its satellite virus (SPMV). We reported AS of defense-related genes including receptor-like kinases, NB-LRR proteins and transcription factors. Strikingly, multiple spliceosome components are themselves alternatively spliced during PMV and SPMV infections. Here, we analyzed the temporal splicing patterns of a splicing factor, Bd-SCL33, following infection of Brachypodium with 6 additional viruses in diverse genera. Our results reveal both dynamic and conserved expression patterns of Bd-SCL33 splice variants during virus infection, and implicate Bd-SCL33 function in response to biotic stresses.
Introduction
Alternative splicing (AS) is a co-transcriptional regulatory process that promotes transcriptome and proteome diversity of the cell. AS is essential for plant development and enhances adaptation to biotic and abiotic stresses.Citation1-4 Using an RNA-seq approach, we recently characterized the AS landscapes of Brachypodium distachyon (Brachypodium), a model grass species,Citation5 infected with Panicum mosaic virus (PMV) and its satellite virus (SPMV).Citation6 We identified virus-triggered AS events of numerous genes involved in plant defenses, as well as transcriptional and post-transcriptional regulatory processes.
AS occurs in the nucleus and requires the activity of several spliceosomal constituents including splicing factors and other ribonucleoproteins.Citation7 Notably, precursor mRNAs (pre-mRNAs) of spliceosomal components themselves are alternatively spliced, often auto-regulating their AS, perhaps to maintain splicing homeostasis.Citation1,8 For example, in Arabidopsis, splicing factor, At-SCL33 undergoes AS during abiotic stresses and hormone treatments.Citation8,9 The At-SCL33 protein also auto-regulates AS of its pre-mRNA by binding to the exon enhancer signals present in the introns.Citation8 We discovered a Brachypodium ortholog of SCL33 (Bd-SCL33) whose splicing patterns are reminiscent of At-SCL33. The expression of Bd-SCL33 splice variants is modulated during plant development, as well as during PMV and PMV+SPMV infections.Citation6 Several Bd-SCL33 splice variants contain premature termination codons (PTCs), and encode truncated proteins ().Citation6 It is possible that virus-induced changes in AS of Bd-SCL33 mediate the genome-wide AS patterns and homeostasis during virus infection.
Table 1. Sequence composition of Bd-SCL33 splice variants in Brachypodiuma
We previously established Brachypodium as a model for diverse host:virus pathosystems.Citation10-12 In addition to PMV and SPMV, Brachypodium is a host for at least 6 additional economically important monocot-infecting viruses.Citation10-12 Here, we further analyzed whether the altered Bd-SCL33 AS patterns observed during PMV and PMV+SPMV infections are also conserved among diverse virus infections in Brachypodium.
Differential splicing of Bd-SCL33 in diverse grass virus infections
Multiple intron-retained Bd-SCL33 splice variants accumulated early during PMV and PMV+SPMV infections ().Citation6 To determine if diverse viruses also modulate Bd-SCL33 AS patterns, we analyzed the temporal expression patterns of Bd-SCL33 splice variants in 6 additional viral infections in Brachypodium (). These grass-infecting viruses included Brome mosaic virus (BMV, genus: Bromovirus), Barley stripe mosaic virus (BSMV, genus: Hordeivirus), Maize mild mottle virus (MMMV, genus: putative Panicovirus), Sorghum yellow banding virus (SYBV, genus: unknown), Wheat streak mosaic virus (WSMV, genus: Tritimovirus), and Foxtail mosaic virus (FoMV, genus: Potexvirus).Citation10 Infected samples were collected at 10, 21, and 42 days post-inoculation (dpi). BMV-, MMMV-, WSMV-, and FoMV-infected plants did not survive through the 42 dpi stage, thus only 10 and 21 dpi stages were analyzed.Citation10 Total RNA from the different samples was isolated using the Direct-zol RNA MiniPrep kit (Zymo Research) and subjected to reverse transcription PCR, as described previously.Citation6
Figure 1. Bd-SCL33 differential splicing during diverse grass virus infections. Expression of Bd-SCL33 transcripts in mock, PMV+SPMV-, BMV-, BSMV-, MMMV-, SYBV-, WSMV- and FoMV-infected Brachypodium at 10, 21 and 42 days postinoculation (dpi), as determined by RT-PCR. The alternatively spliced Bd-SCL33 transcripts, as defined in , are identified by an asterisk (*). UBC18 was used as control for approximately equal cDNA amounts in the different samples. Lane “M” indicates DNA molecular weight markers.
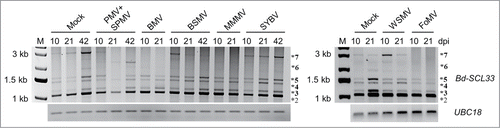
In Brachypodium, expression of the Bd-SCL33 primary transcript, Bradi1g61330.1 (, #1), was below detectable levels, while expression of AS transcript, Bradi1g61330.2 (, #2) was predominant throughout the developmental stages . Expression of the intron-retained Bd-SCL33 splice variants (#3, #4, #5 and #7) was developmentally regulated ().Citation6 For example, in mock-inoculated plants, levels of these splice variants accumulated as the plants matured (21 and 42 dpi) (), further supporting our earlier observations.Citation6 Interestingly, the developmental expression pattern of these splice variants was perturbed during most virus infections, with the exception of SYBV and WSMV (). In a manner similar to PMV and PMV+SPMV infections,Citation6 multiple viruses triggered an earlier accumulation of splice variants #3, #4 and #5, as early as 10 dpi, compared to mock-inoculated plants (). The abundance of splice variant #7, representing the full-length pre-mRNA of Bd-SCL33, was similarly induced in BSMV-, SYBV-, and WSMV-infected individuals at 10 dpi, compared to mock. In addition to the conserved Bd-SCL33 AS patterns, we also found unique patterns. For example, abundance of splice variant #6, an intron I and intron III retained isoform (), is induced in PMV+SPMV infection,Citation6 but not in the remaining 6 virus infections ().
Conclusion
Our findings demonstrate that diverse grass virus infections trigger conserved and unique changes in Bd-SCL33 alternative splicing. The Bd-SCL33 splice variants contain pre-mature termination codons and encode truncated proteins, retaining only partial regions of the N-terminal RNA recognition motif (RRM) domain ().Citation6 It is possible that the truncated proteins inhibit the activity of full-length SCL33 protein in a dominant negative manner. Such modulation of SCL33 activity, and perhaps of the other splicing regulators during virus infection,Citation6 could represent a means to control genome-wide changes in AS landscapes during biotic stress. Whether the AS changes occurring during infection promote plant adaptation to virus stress or contributes to virulence of the viruses remains to be investigated.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We appreciate the comments of Sonia Irigoyen (Texas A&M AgriLife) during the preparation of this manuscript.
Funding
This study was partially supported by funds from Texas A&M AgriLife Research Start-up Fund to K.K.M.
References
- Duque P. A role for SR proteins in plant stress responses. Plant Signal Behav 2011; 6:49-54
- Reddy ASN, Marquez Y, Kalyna M, Barta A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013; 25:3657-83; PMID:24179125; http://dx.doi.org/10.1105/tpc.113.117523
- Staiger D, Brown JWS. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013; 25:3640-56; PMID:24179132; http://dx.doi.org/10.1105/tpc.113.113803
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010; 463:457-63; PMID:20110989; http://dx.doi.org/10.1038/nature08909
- Brkljacic J, Grotewold E, Scholl R, Mockler T, Garvin DF, Vain P, Brutnell T, Sibout R, Bevan M, Budak H, et al. Brachypodium as a model for the grasses: today and the future. Plant Physiol 2011; 157:3-13; PMID:21771916; http://dx.doi.org/10.1104/pp.111.179531
- Mandadi KK, Scholthof K-BG. Genome-wide analysis of alternative splicing landscapes modulated during plant-virus interactions in Brachypodium distachyon. Plant Cell 2015; 27:71-85; PMID:25634987; http://dx.doi.org/10.1105/tpc.114.133991
- Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol 2011; 3; pii: a003707; PMID:21441581
- Thomas J, Palusa SG, Prasad KVSK, Ali GS, Surabhi G-K, Ben-Hur A, Abdel-Ghany SE, Reddy AS. Identification of an intronic splicing regulatory element involved in auto-regulation of alternative splicing of SCL33 pre-mRNA. Plant J 2012; 72:935-46; PMID:22913769
- Thomas J. Functional analysis of three Arabidopsis SR proteins (SCL33, SC35, SCL30A) in plant development and splicing. PhD dissertation (Fort Collins: Colorado State University, 2012)
- Mandadi KK, Pyle JD, Scholthof K-BG. Comparative analysis of antiviral responses in Brachypodium distachyon and Setaria viridis reveals conserved and unique outcomes among C3 and C4 plant defenses. Mol Plant Microbe Interact 2014; 27:1277-90
- Mandadi KK, Scholthof K-BG. Characterization of a viral synergism in the monocot Brachypodium distachyon reveals distinctly altered host molecular processes associated with disease. Plant Physiol 2012; 160:1432-52
- Mandadi KK, Scholthof K-BG. Plant immune responses against viruses: how does a virus cause disease? Plant Cell 2013; 25:1489-505; PMID:23709626; http://dx.doi.org/10.1105/tpc.113.111658
