Abstract
miRNAs might move cell to cell and act as mobile signals in plant development, while the regulatory mechanisms of miRNA cell-to-cell movement are still unclear. Recently, in Arabidopsis leaf primordia, we revealed that miR165 from the MIR165A gene, which is expressed in the abaxial epidermal cells of leaf primordia, acts non-cell-autonomously in inner cells on the abaxial side. We proposed that not only mature miR165 sequence but also the MIR165A primary transcript sequence are required for the confinement of miR165 activity to the abaxial side of leaf primordia. The deletion analysis of the MIR165A genomic fragment showed that with a lack of the 3' region of MIR165A its activity is not confined in leaf primordia, suggesting that the full-length primary transcript of MIR165A is important for the regulatory mechanism of miRNA activity confinement in leaf primordia. It has been reported that the MIR165A transcript is predicted to be translated into the short poly peptide, proposing that the MIR165A transcript may be exported to the cytoplasm. Considering these matters, we propose a hypothesis for the confinement of miR165 activity to the abaxial side in leaf primordia dependent on the MIR165A primary transcript.
Abbreviations
| DCL1 | = | DICER-LIKE1 |
| HD-Zip III | = | class III homeodomain-leucine zipper |
| MIRNA | = | MICRORNA |
| miRNA | = | microRNA |
| MIR165/166 | = | MICRORNA165/166 |
| MIR165A | = | MICRORNA165A |
| miR165/166 | = | microRNA165/166 |
| miR390 | = | microRNA390 |
| miR399 | = | microRNA399 |
| miPEP | = | miRNA-encoded peptide |
| miPEP165a | = | pri-miR165a encoded peptide. |
Small RNAs, including miRNAs, can repress the expression of their target genes in a non-cell-autonomous manner. A miRNA, miR399, whose precursor was expressed in the shoot, might move from shoot to root via phloem and represses the expression of its target gene.Citation1 In maize, precursor transcripts of miR390, which is a regulatory factor for the determination of the abaxial side in leaf polarity, accumulate only in the L1 cell layer of the SAM but mature miR390 is detected in both the L1 and L2 layers of leaf primordia.Citation2 In Arabidopsis roots, miR165/166-encoding genes, MIR165A, MIR166A and MIR166B, are expressed in the endodermis and miR165/166 is likely to move to the stele to repress HD-Zip III expression.Citation3,4 MIR165/166 genes also show non-cell-autonomous activity during embryogenesis.Citation5 These data indicate that miRNAs are able to act as mobile signals for development. However, although miRNAs might move cell to cell through plasmodesmata,Citation6,7 the regulatory mechanisms for the cell-to-cell movement of miRNAs are still unknown.
During leaf development along the adaxial-abaxial polarity in Arabidopsis thaliana, rigid determination of the expression domain of HD-Zip III is required in leaf primordia.Citation8,9 The leaf primordia have 6 cell layers along the adaxial-abaxial axis; adaxial epidermis, adaxial L2, adaxial L3, abaxial L3, abaxial L2 and abaxial epidermis. () In our recent publication,Citation10 we reported that MIR165A, which is expressed in the abaxial epidermis of leaf primordia, clearly represses the expression of HD-Zip III genes in 3 cell layers in the abaxial side but not those in the adaxial side, indicating that MIR165A is able to act in non-cell autonomous and abaxial-baiased manners in leaf primordia.() In order to analyze the activity pattern of MIR165A in leaf primordia without any effects of other MIR165/166 genes, we used mutated miR165 (miR165mu) having 2 nucleotide mutation in miR165 sequence of the MIR165A gene, because miR165mu do not have any effects on the leaf development but show the same activity pattern as that of native miR165/166. We also suggested that the movement of miR165 arisen from MIR165A is restricted to within a few cells toward to the adaxial side in the leaf primordia, confining miR165 activity to the abaxial side. In addition, we found that both a mature miR165 sequence and the MIR165A backbone sequence are required to confine miR165 activity to the abaxial side in leaf primordia.
Figure 1. (A) Primary transcripts of MIR165A and 3' end-truncated MIR165A. Red and orange boxes indicates miR165 and miR165a*, respectively. Gray boxes represent pri-miR165a. A dashed-line box indicate the lack of the 3'-region in 3' end-truncated MIR165A. (B) Schematic model of the activity patterns of miR165 arisen from the MIR165A (left) and 3' end-truncated MIR165A (right) in leaf primordia. Black color indicates the MIR165A-expressing and miR165-active cell layer, and gray color represents miR165-active cell layers.
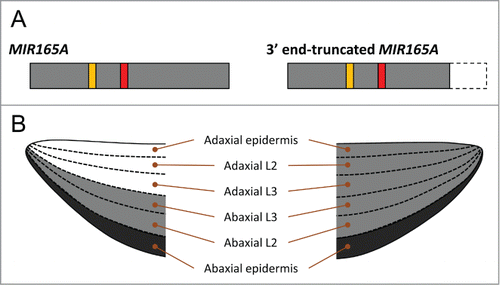
At the same time, we also revealed another regulatory element in the MIR165A genomic region that confines the miR165 activity. When miR165mu was expressed by either the MIR165Amu(+241) or MIR165Amu(+365) transgenes, both of which lack the 3′ region of the untranscribed sequence of MIR165A, the activity pattern of miR165mu was very complicated; some plants showed the miR165mu activity throughout the leaf primordia, while other plants showed the activity only on the abaxial side (see Supporting information of Tatematsu et al.Citation10) The results suggest that the 3' untranscribed region of the MIR165A gene is required for the stable confinement of miR165 activity to the abaxial side in leaf primordia. Yao et al.Citation11 analyzed the transgenic lines, including p165a:pri-miR165a-GUS, which expresses the 3' end-truncated MIR165A primary transcript (pri-miR165a) fused to the GUS gene. Some of the p165a:pri-miR165a-GUS transgenic plants exhibited either single radial-symmetric or severely up-curled cotyledons and leaves, suggesting that pri-miR165a-GUS expressed in the abaxial epidermis represses HD-Zip III expression even on the adaxial side of the leaves, which cause the abaxialzed leaf phenotype. This result also suggests that the 3' end-truncated pri-miR165a fused to the GUS sequence cannot confine the miR165 activity in leaf primordia. Considering that the nucleotide sequence directly after the 3′ termination site is required for the proper termination of transcription, it is possible that aberrant pri-miR165a, either shorter transcripts or longer transcripts with a binary vector sequence, were transcribed from MIR165Amu(+241) and MIR165Amu(+365), and that the aberrant transcripts cannot confine the mobility of miR165mu in leaf primordia. () Since the aberrant pri-miR165a transcripts would have full length of pre-miR165a, which include miR165, loop, and miR165a*, the entire sequence of MIR165A primary transcript is important for the confinement of its activity.
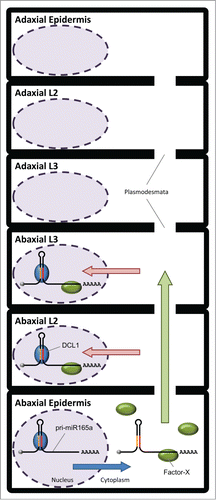
How does the primary transcript sequence of MIR165A contribute on the confinement of its activity to the abaxial side of leaf primordia? The primary transcripts of MIRNA genes have been thought to be localized mainly in nucleus and are processed by DCL1. Recently, Lauressergues and co-authorsCitation12 reported that some MIRNA primary transcripts encode short peptides, call miPEPs, and that these miPEPs have a role in enhancing transcription of their own primary transcripts. They also showed that the pri-miR165a is predicted to be translated into miPEP165a, which is composed of 18 amino acid residues, proposing that the pri-miR165a is exported form nucleus to cytoplasm. Taken together with the requirement of the full-length sequence of pri-miR165a, we hypothesize a possible scenario to confine miR165 activity pattern to the abaxial side, which depends on the primary transcript of the MIR165A, in Arabidopsis leaf primordia. () Firstly, in the abaxial epidermal cells, a part of pri-miR165a is exported from the nucleus to the cytoplasm after transcription from the MIR165A gene. An unknown factor, Factor-X, is thought to bind to the 3′ end-region of the pri-miR165a to form a pri-miR165a/Factor-X complex. Secondly, the pri-miR165a/Factor-X complex might be allowed to move to within a few cells toward to the adaxial side via plasmodesmata but not to reach the cells on the adaxial side. Thirdly, in the abaxial L2 and L3 cells, the pri-miR165a/Factor-X complex is imported into the nucleus. Finally, the pri-miR165a is processed by DCL1, and mature miR165 represses the expression of HD-Zip III in the abaxial-side cells.
Figure 2. Schematic model of a possible scenario for the confinement of miR165 activity pattern dependent on the MIR165A primary transcript (pri-miR165a) in leaf primordia. In the abaxial epidermal cells, a part of pri-miR165a is exported from the nucleus to the cytoplasm (blue arrow), and Factor-X binds to the 3' end-region of the pri-miR165a. The pri-miR165a/Factor-X complex moves to the abaxial L2 and L3 cells through plasmodesmata (green arrow). In the abaxial L2 and L3 cells, the pri-miR165a/Factor-X complex is imported into the nucleus (red arrow), and the pri-miR165a is dissected by DCL1 in the nucleus.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors acknowledge Mr. Colin Betha Cecil (National Institute for Basic Biology) for critical reading of this manuscript.
References
- Pant BD, Buhtz A, Kehr J, Scheible WR. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 2008; 53:731-8; PMID:17988220; http://dx.doi.org/10.1111/j.1365-313X.2007.03363.x
- Nogueira FTS, Chitwood DH, Madi S, Ohtsu K, Schnable PS, Scanlon MJ, Timmermans MCP. Regulation of small RNA accumulation in the maize shoot apex. Plos Genet 2009; 5:e1000320; PMID:19119413; http://dx.doi.org/10.1371/journal.pgen.1000320
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vaten A, Thitamadee S, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 2010; 465:316-21; PMID:20410882; http://dx.doi.org/10.1038/nature08977
- Miyashima S, Koi S, Hashimoto T, Nakajima K. Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 2011; 138:2303-13; PMID:21558378; http://dx.doi.org/10.1242/dev.060491
- Miyashima S, Honda M, Hashimoto K, Tatematsu K, Hashimoto T, Sato-Nara K, Okada K, Nakajima K. A Comprehensive expression analysis of the Arabidopsis micoRNA165/6 gene family during embryogenesis reveals a conserved role in meristem specification and a non-cell-autonomous function. Plant Cell Physiol 2013; 54:375-84; PMID:23292599; http://dx.doi.org/10.1093/pcp/pcs188
- Hyun TK, Uddin MN, Rim Y, Kim JY. Cell-to-cell trafficking of RNA and RNA silencing through plasmodesmata. Protoplasma 2011; 248:101-16; PMID:21042816; http://dx.doi.org/10.1007/s00709-010-0225-6
- Vaten A, Dettmer J, Wu S, Stierhof YD, Miyashima S, Yadav SR, Roberts CJ, Campilho A, Bulone V, Lichtenberger R, et al. Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell 2011; 21:1144-55; PMID:22172675; http://dx.doi.org/10.1016/j.devcel.2011.10.006
- Toyokura K, Watanabe K, Oiwaka A, Kusano M, Tameshige T, Tatematsu K, Matsumoto N, Tsugeki R, Saito K, Okada K. Succinic semialdehyde dehydrogenase is involved in the robust patterning of Arabidopsis leaves along the adaxial-abaxial axis. Plant Cell Physiol 2011; 52:1340-53; PMID:21690177; http://dx.doi.org/10.1093/pcp/pcr079
- Tameshige T, Fujita H, Watanabe K, Toyokura K, Kondo M, Tatematsu K, Matsumoto N, Tsugeki R, Kawaguchi M, Nishimura M, et al. Pattern dynamics in adaxial-abaxial specific gene expression are modulated by a plastid retrograde signal during Arabidopsis thaliana leaf development. Plos Genet 2013; 9:e1003655; PMID:23935517; http://dx.doi.org/10.1371/journal.pgen.1003655
- Tatematsu K, Toyokura K, Miyashima S, Nakajima K, Okada K. A molecular mechanism that confines the activity pattern of miR165 in Arabidopsis leaf primordia. Plant J 2015; 82:596-608; PMID:25788175; http://dx.doi.org/10.1111/tpj.12834
- Yao XZ, Wang H, Li H, Yuan ZH, Li FP, Yang L, Huang H. Two types of cis-acting elements control the abaxial epidermis-specific transcription of the MIR165a and MIR166a genes. FEBS Lett 2009; 583:3711-7; PMID:19879265; http://dx.doi.org/10.1016/j.febslet.2009.10.076
- Lauressergues D, Couzigou JM, Clemente HS, Martinez Y, Dunand C, Bécard G, Combier JP. Primary transcripts of microRNAs encode regulatory peptide. Nature 2015; 520:90-3; PMID:25807486; http://dx.doi.org/10.1038/nature14346
