ABSTRACT
Legume plants have developed the capacity to establish symbiotic interactions with soil bacteria (known as rhizobia) that can convert N2 to molecular forms that are incorporated into the plant metabolism. The first step of this relationship is the recognition of bacteria by the plant, which allows to distinguish potentially harmful species from symbiotic partners. The main molecular determinant of this symbiotic interaction is the Nod Factor, a diffusible lipochitooligosaccharide molecule produced by rhizobia and perceived by LysM receptor kinases; however, other important molecules involved in the specific recognition have emerged over the years. Secreted exopolysaccharides and the lipopolysaccharides present in the bacterial cell wall have been proposed to act as signaling molecules, triggering the expression of specific genes related to the symbiotic process. In this review we will briefly discuss how transcriptomic analysis are helping to understand how multiple signaling pathways, triggered by the perception of different molecules produced by rhizobia, control the genetic programs of root nodule organogenesis and bacterial infection. This knowledge can help to understand how legumes have evolved to recognize and establish complex ecological relationships with particular species and strains of rhizobia, adjusting gene expression in response to identity determinants of bacteria.
Introduction
Plants constantly need to discriminate between organisms that are potentially harmful from those that can have a positive impact in their lives. One of the most relevant questions in terms of ecological interactions is how plants recognize and discriminate between pathogenic and mutualistic microorganisms present in their environment. In the last years, important progresses in the understanding of the molecular mechanisms involved in the recognition of bacteria and the signals involved have been achieved. However, some important questions remain open and are the subject of intense research. In this review we will describe the current knowledge on the role of main signal molecules in the legume-rhizobia symbiosis and discuss how transcriptomic analysis has helped to dissect the role played by the different components of the molecular dialog between legume plants and bacteria.
Opening the gate
In the presence of compatible rhizobia, roots of legume plants initiate an infection program that facilitates the penetration of bacteria into the nodule, where they will be part of the nitrogen-fixing structures called symbiosomes. In the most sophisticated mode of infection mechanisms, a tubular structure called infection thread connects the epidermis with internal cell layers of the root; therefore, recognition of the right rhizobia is critical to avoid pathogen or opportunistic infections. This type of infection requires the activation of a genetic program in the plant root that must be triggered in the right place, at the right moment. There are several species from different genera of bacteria that can colonize roots of legume plants. The specificity of the interaction is determined by the exchange of molecules between both symbiotic partners, establishing a narrow range of legumes that can be infected by each rhizobial species. There are some exceptions, like Rhizobium sp. NGR234, which is able to nodulate a broad range of legume plants.Citation1 Besides this first check point, there are some examples of plants that are able to discriminate biovars or strains of rhizobia that nodulate more efficiently.Citation2-3 Research progress made in the last years have positioned the specific interaction between bacterial signal molecules and receptor-like kinases (RLKs) present in the epidermis of roots in the center of the plant decision to either trigger a defense response to avoid the entrance of pathogens or, alternatively, suppress the defense response to allow infection by symbiotic bacteria.()
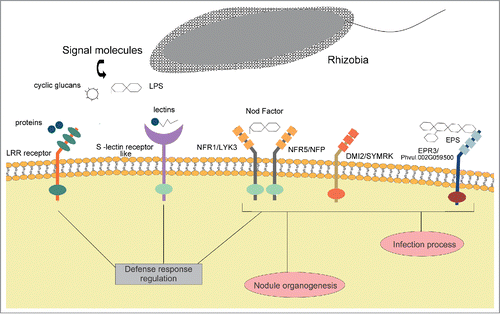
Figure 1. Rhizobia perception mediated by plant receptors. Nod Factor (NF) perception is required for infection and nodule organogenesis, but other signaling molecules secreted by bacteria, such as exopolysaccharides (EPS), proteins, cyclic glucans and K antigens and cell wall associated molecules, like the lipopolysaccharide (LPS) have been demonstrated to modulate the infection process. NF and EPS are recognized by LysM receptor-like kinases (LysM RLKs). In particular, NF is perceived by a receptor complex comprising the LysM receptors NFP/LYK3 in M. truncatula or NFR1/NFR5 in L. japonicus. NF signaling activates the root nodule symbiotic pathway and inhibits defense responses. DMI2 and SYMRK are receptors from M. truncatula and L. japonicus that are required for nodulation, but their ligands are unknown. The EPS receptor (EPR3 in L. japonicus, Phvul.002G059500 in common bean) plays a major role in the infection process, suggesting a sequential receptor mediated recognition of NF and EPS. Rhizobial proteins and lectins are recognized by LRR and lectin RLKs, respectively, and have been proposed to regulate defense responses. EPS and LPS negatively regulate two receptors with serine/threonine (Ser/Thr) kinase domains highly similar to the ethylene receptor ETR2 by an unknown process.Citation31 These receptors are regulated by abiotic stresses and hormones. Abbreviations: LRR: leucine rich repeats; NFR: Nod factor receptor; NFP: Nod factor perception; LYK3: LysM receptor kinase 3; DMI2: Does not Make Infection; SYMRK: Symbiosis receptor-like kinase; EPR3: EPS receptor 3; LysM: Lysin motif.
Nod factor, the key of specificity
When legume plants are in the need of nitrogen, their roots produce and secrete flavonoids/isoflavonoids to the rhizosphere. Rhizobia sense these molecules and activate the product of the nodD gene, a transcription factor that controls the expression of genes involved in the synthesis of the Nod Factor (NF). NFs are secreted lipochitooligosaccharides (LCO) whose chemical modifications depend on the rhizobium species that produce them and act as the main determinants of host specificity (). The biological role of LCO molecules has been recently reviewed.Citation4 They act at nanomolar concentrations, inducing many early molecular and physiological changes occurring in the root hair. NFs are recognized by receptors with LysM extracellular domains. Single mutations in the NF receptors are sufficient to change specificity of the interaction at the species level.Citation5 Interestingly, this family of receptors is also critical in the recognition of fungi during the mutualistic mycorrhiza interaction in Parasponia andersonii.Citation6 The discovery that the mycorhization factor (Myc factor) is also a LCO suggests that the Rhizobium NF perception system evolved from the ancient mycorrhizal symbiosis. RLKs with extracellular LysM domains have been also implicated in the recognition of fungal pathogens. In particular, CERK1 (Chitin Elicitor Receptor Kinase 1) is required for chitin-induced defense responses in Arabidopsis thaliana and rice (Oryza sativa),Citation7-8 suggesting an evolutionary connection between recognition associated to pathogenic and symbiotic interactions.
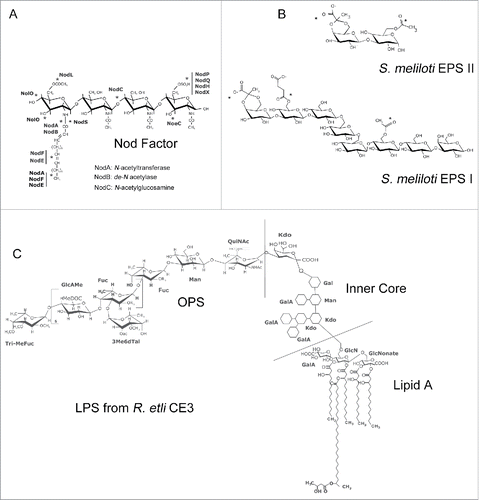
Figure 2. Rhizobium Signal molecules. Representative structures of Nod Factors (NF) exopolysaccarides (EPS) and lipopolysaccharides (LPS). (A) The typical NF backbone consists of 4 or 5 β-1-4 linked N-acetyl-glucosamine residues. NFs are subject to chemical modifications (the position of frequently added groups are indicated with asterisks) by the action of rhizobia nod genes (in bold). The different types of decorations result in a mix of NFs produced by each species of rhizobia. The product of the nodA, nodB and nodC genes participate in the synthesis of the NF backbone. (B) The EPS molecules from S. meliloti Rm1021 are EPS II and EPS I. EPS II is a galactoglucan molecule, whereas EPS I consists of repeating units of octasaccharides modified with acetyl, succinyl and pyruvyl substituents (indicated with asterisks) and is also known as succinoglycan (C) Chemical structure of the LPS from R. etli CE3. LPS is constituted by 3 modules: lipid A, an inner core oligosaccharide and a highly variable O-antigen polysaccharide (OPS). LPSs from rhizobia have variable OPS regions and a number of unique characteristics compared with LPS from enteric bacterial species. The OPS and lipid A regions are key components of the legume-rhizobia interaction.
Cell wall determinants of rhizobial identity
The identity of the bacteria can also be determined by the presence of surface molecules present in the cell wall that can be recognized by surface receptors from the plant. Several glycans have been shown to play a role in nodulation, such as exopolysaccharides (EPS), lipopolysaccharides (LPS), cyclic glucans and capsular polysaccharides.
EPS, a second stage of recognition
Multiple roles have been proposed for EPS as a signal molecule, from an active function during infection Citation9-11 to a suppression of defense responses.Citation12-13 The composition of EPS differs among rhizobial species, but also among strains, suggesting it can act as a determinant of specificity. The structure and biosynthesis of EPS have been studied in detail in Sinorhizobium meliloti (). In addition, several mutants of Mesorrhizobium loti were used to characterize the requirements of EPS during early stages of the interaction, showing that defects in the biosynthesis of EPS can affect different stages of the organogenesis of determinate nodules.Citation14 Using one of these strains, Kawaharada and collaborators identified a LysM RLK with an extracellular domain able to bind the M. loti EPS.Citation15 Expression of this receptor is inducible by NF and sufficient to select compatible bacteria through the recognition of the EPS. This work supports the hypothesis of a two-stage mechanism involving recognition of both NF and EPS to sustain infection inside the root hair.
LPS, suppression of the immune responses
LPS is composed by an oligosaccharide and a lipid that anchor the molecule to the outer membrane of the gram-negative bacteria (). An O-antigen saccharide polymer is attached to the inner core of the molecule and exposed on the surface on the bacterial cell wall, where the interaction with the plant is critical. Mutants in the synthesis of LPS are often affected in the infection, producing phenotypes similar to those in which defense reactions are triggered.Citation16-18 In addition, exogenous application of LPS suppresses the oxidative burst reaction in alfalfa (Medicago sativa) Citation19 or in M. truncatula roots.Citation20 These results led to the proposal that LPS could act as a suppressor of defense responses, a key step to sustain the progression of bacteria inside the infection thread. However, the effect of LPS in nodulation varies in different species.Citation21-22
All together, these results point to the existence of multiple molecules that participate in the initial recognition of bacteria by plant receptors, attachment to the surface of root hairs, progression of infection structures and suppression of the plant immune response. The action of these molecules depends on the molecular structure and their effects can be differently exerted depending on the host plant, suggesting an active role as signaling molecules.
Transcriptomic studies with mutant strains
The study of differences in gene expression have provided valuable information to understand key biological processes. Array-based technologies were followed by the development of RNA sequencing (RNA-seq), allowing characterization of transcriptional changes at a global scale. Several studies have contributed to understand transcriptional reprogramming during symbiosis in the model legumes M. truncatula and Lotus japonicus.Citation23-30 Genes involved in cytoskeleton rearrangements, cell cycle reinitiation, transcriptional activation, signaling and perception, as well as hundreds of genes with unknown function, have been identified as differentially modulated at different stages of the symbiotic process. RNA-seq analysis, unlike array-based transcriptomic, also contributed to the discovery of new genes and mRNA processing variants in the context of nodulation. As an example, transcripts originated from alternative processing (different transcription start sites or alternative splicing) were identified as differentially accumulated during the early stages of the interaction between common bean (Phaseolus vulgaris) and Rhizobium etli.Citation31
In order to validate the role of NF, EPS and LPS as signal molecules and to characterize the genes that are modulated by the presence of these determinants in the bacterial environment, transcriptomic studies have provided insights on the putative roles of the specific recognition of these molecules. Of particular interest are studies that used mutant strains of rhizobia to dissect the transcriptional changes associated to recognition of signal molecules from bacteria.Citation28,30-33 (). Jones and collaborators Citation32 showed the differences in the M. truncatula transcriptional response when it was inoculated with the wild type strain or a succinoglycan-deficient strain of S. meliloti unable to establish a successful infection. The absence of this exopolysaccharide resulted in a reduction of genes involved in protein synthesis and degradation, as well as genes involved in nodulation (known as nodulins). On the other hand, defense response genes and a set of unknown genes were up-regulated in response to the EPS-deficient mutant, suggesting that recognition of the EPS is required to initiate the infection process, but also to suppress defense responses. A similar study using the exoA mutant strain of S. meliloti identified a group of genes that are induced in immature nodules (4 days after inoculation) in response to the wild type strain in the infection zone of the nodule, but not in response to the exoA strain. Some of these genes seems to be involved in meristem activity and differentiation, including genes that participate in cytokinin synthesis and activation.Citation30 At 10 days after inoculation, a group of plant defense genes showed higher mRNA levels in plants inoculated with exoA, although they were still expressed in response to the wild type strain of S. meliloti, providing partial support to the proposed role of EPS in the limitation of plant defense responses.
Table 1. Transcriptomic analysis of legumes infected with mutant bacterial strains.
mRNA levels of genes involved in plant defense are high 1 hour after inoculation of M. truncatula plants with the S. meliloti wild type strain, but decrease at 6 hours post-infection,Citation25 leading to the idea that defense responses were transiently induced and then suppressed concomitantly with progression of the infection process. As previously mentioned, the LPS has been proposed as a suppressor of this early defense response. Transcriptomic studies using LPS mutant strains have been conducted at different times and in different biological systems, making difficult to test this hypothesis. For example, D'Antuono and collaborators Citation33 used a strain of M. loti with reduced levels of LPS (lpsβ2) that was able to form normal nodules and characterized transcriptional changes in L. japonicus by microarray analysis at 7 and 28 days after inoculation. Some of the defense responses induced in developing nodules formed with the wild type strain were not present when the lpsβ2 strains was used. Later on, Maunoury at al Citation28 studied the lspB mutant strain of S. meliloti, which is able to differentiate to bacteroids inside the nodules. The effect of this mutant in the transcriptome of M. truncatula was similar to the wild type strain according to a Principal Component Analysis, indicating that the nodules formed have already overpassed the early defense reactions triggered by the plant. More recently, we have studied the transcriptional changes of common bean in response to LPS and EPS mutant strains of R. etli.Citation31 In this system, the LPS mutant did not form nodules, whereas the EPS strain showed a phenotype similar to the wild type. At 24 hours post inoculation, defense responses were still active in roots inoculated with the wild type strain, but no significant effects of LPS and EPS were detected at this early time point of the interaction. In addition, this work highlights novel aspects of the responses regulated by NF, EPS and LPS, as the regulation of circadian rhythms at early stages of the interaction and genes that participate in the transcriptional and posttranscriptional silencing mediated by small RNAs.
Future perspectives
Understanding the molecular mechanisms involved in the plant recognition of microorganisms will be the key to manipulate ecological relationships in the context of agricultural systems. It is a common practice to add inoculants to the soil to improve plant growing or to substitute the usage of chemically synthesized organic fertilizers. In order to get the desire effect, it is critical that plants recognize and establish the right type of interaction with the microorganisms present in their environment. The selection of the right surface determinants can be the key to improve the competition capacity, preventing other bacteria already present in the soil to displace strains of bacteria added as inoculants, which are more efficient in terms of nitrogen fixation. Transcriptional studies can help to understand how plants respond to determinants of bacteria identity, triggering genetic programs that will determine the ecological relationship between these organisms. At the same time, they provide the basis to select genes for functional analysis, but, in order to extract solid conclusions, it is critical to understand the particular aspects of each biological interaction and to select the stages of the interaction to perform the study accordingly.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was financially supported by grants from ANPCyT, Argentina (PICT 2010/2722 and 2010/2431). MEZ and FAB are members of CONICET
References
- Pueppke SG, Broughton WJ. Rhizobium sp strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol Plant Microbe Interact 1999; 12:293-318; PMID:10188270; http://dx.doi.org/10.1094/MPMI.1999.12.4.293
- Aguilar OM, Riva O, Peltzer E. Analysis of Rhizobium etli and of its symbiosis with wild Phaseolus vulgaris supports coevolution in centers of host diversification. Proc Natl Acad Sci USA 2004; 101:13548-53; PMID:15340138; http://dx.doi.org/10.1073/pnas.0405321101
- Yang S, Tang F, Gao M, Krishnan HB, Zhu H. R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc Natl Acad Sci USA 2010; 107:18735-40; PMID:20937853; http://dx.doi.org/10.1073/pnas.1011957107
- Limpens E, Zeijl AV, Geurts R. Lipochitooligosaccharides modulate plant host immunity to enable endosymbioses. Annu Rev Phytopathol 2015; 53:311-34; PMID:26047562; http://dx.doi.org/10.1146/annurev-phyto-080614-120149
- Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EM, Albrektsen AS, James EK, Thirup S, Stougaard J. LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J 2007; 26:3923-35; PMID:17690687; http://dx.doi.org/10.1038/sj.emboj.7601826
- Maillet F, Poinsot V, Andre O, Puech-Pages V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 2011; 469:58-63; PMID:21209659; http://dx.doi.org/10.1038/nature09622
- Petutschnig EK, Jones AME, Serazetdinova L, Lipka U, Lipka V. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced posphorylation. J Biol Chem 2010; 285:28902-11; PMID:20610395; http://dx.doi.org/10.1074/jbc.M110.116657
- Shinya T, Motoyama N, Ikeda A, Wada M, Kamiya K, Hayafune M, Kaku H, Shibuya N. Functional characterization of CEBiP and CERK1 homologs in Arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol 2012; 53:1696-706; PMID:22891159; http://dx.doi.org/10.1093/pcp/pcs113
- Cheng HP, Walker GC. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol 1998; 180:5183-91; PMID:9748453
- Laus MC, Logman TJ, van Brussel AAN, Carlson RW, Azadi P, Gao MY, Kijne JW. Involvement of exo5 in production of surface polysaccharides in Rhizobium leguminosarum and its role in nodulation of Vicia sativa subsp. nigra. J Bacteriol 2004; 186:6617-25; PMID:15375143; http://dx.doi.org/10.1128/JB.186.19.6617-6625.2004
- Pellock BJ, Cheng HP, Walker GC. Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti polysaccharides. J Bacteriol 2000; 182:4310-8; PMID:10894742; http://dx.doi.org/10.1128/JB.182.15.4310-4318.2000
- Niehaus K, Kapp D, Pühler A. Plant defence and delayed infection of alfalfa pseudonodules induced by an exopolysaccharide (EPS I)-deficient Rhizobium meliloti mutant. Planta 1993; 190:415-25; http://dx.doi.org/10.1007/BF00196971
- Parniske M, Schmidt P, Kosch K, Muller P. Plant defense responses of host plants with determinate nodules induced by EPS-defective exoB mutants of Bradyrhizobium japonicum. Mol Plant Microbe Interact 1994; 7:631-8; http://dx.doi.org/10.1094/MPMI-7-0631
- Kelly SJ, Muszyński A, Kawaharada Y, Hubber AM, Sullivan JT, Sandal N, Carlson RW, Stougaard J, Ronson CW. Conditional requirement for exopolysaccharide in the Mesorhizobium–Lotus symbiosis. Mol Plant Microbe Interact 2012; 26:319-29; PMID:23134480; http://dx.doi.org/10.1094/MPMI-09-12-0227-R
- Kawaharada Y, Kelly S, Nielsen MW, Hjuler CT, Gysel K, Muszynski A, Carlson RW, Thygesen MB, Sandal N, Asmussen MH, et al. Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 2015; 523:308-12; PMID:26153863; http://dx.doi.org/10.1038/nature14611
- Perotto S, Brewin N, Kannenberg E. Cytological evidence for a host defense response that reduces cell and tissue invasion in pea nodules by lipopolysaccharide-defective mutants of Rhizobium leguminosarum strain 3841 Mol Plant Microbe Interact 1994; 7:99-112; http://dx.doi.org/10.1094/MPMI-7-0099
- García-de los Santos A, Brom S. Characterization of two plasmid-borne lpsβ loci of Rhizobium etli required for lipopolysaccharide synthesis and for optimal interaction with plants. Mol Plant Microbe Interact 1997; 10:891-902; PMID:9304861; http://dx.doi.org/10.1094/MPMI.1997.10.7.891
- Gao M, D'Haeze W, De Rycke R, Wolucka B, Holsters M. Knockout of an azorhizobial dTDP-L-rhamnose synthase affects lipopolysaccharide and extracellular polysaccharide production and disables symbiosis with Sesbania rostrata. Mol Plant Microbe Interact 2001; 14:857-66; PMID:11437259; http://dx.doi.org/10.1094/MPMI.2001.14.7.857
- Albus U, Baier R, Holst O, Pühler A, Niehaus K. Suppression of an elicitor-induced oxidative burst reaction in Medicago sativa cell cultures by Sinorhizobium meliloti lipopolysaccharides. New Phytol 2001; 151:597-606; http://dx.doi.org/10.1046/j.0028-646x.2001.00214.x
- Scheidle H, Groß A, Niehaus K. The Lipid A substructure of the Sinorhizobium meliloti lipopolysaccharides is sufficient to suppress the oxidative burst in host plants. New Phytol 2005; 165:559-66; PMID:15720666; http://dx.doi.org/10.1111/j.1469-8137.2004.01214.x
- Niehaus K, Lagares A, Pühler A. A Sinorhizobium meliloti lipopolysaccharide mutant induces effective nodules on the host plant Medicago sativa (Alfalfa) but fails to establish a symbiosis with Medicago truncatula. Mol Plant Microbe Interact 1998; 11:906-14; http://dx.doi.org/10.1094/MPMI.1998.11.9.906
- Noel KD, Vandenbosch KA, Kulpaca B. Mutations in Rhizobium phaseoli that lead to arrested development of infection threads. J Bacteriol 1986; 168:1392-401; PMID:3782040
- Roux B, Rodde N, Jardinaud MF, Timmers T, Sauviac L, Cottret L, Carrère S, Sallet E, Courcelle E, Moreau S, et al. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J 2014; 77:817-37; PMID:24483147; http://dx.doi.org/10.1111/tpj.12442
- El Yahyaoui F, Küster H, Ben Amor B, Hohnjec N, Pühler A, Becker A, Gouzy J, Vernié T, Gough C, Niebel A, et al. Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol 2004; 136:3159-76; PMID:15466239; http://dx.doi.org/10.1104/pp.104.043612
- Lohar DP, Sharopova N, Endre G, Penuela S, Samac D, Town C, Silverstein KA, VandenBosch KA. Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol 2006; 140:221-34; PMID:16377745; http://dx.doi.org/10.1104/pp.105.070326
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. A gene expression atlas of the model legume Medicago truncatula. Plant J 2008; 55:504-13; PMID:18410479; http://dx.doi.org/10.1111/j.1365-313X.2008.03519.x
- Kouchi H, Shimomura K, Hata S, Hirota A, Wu GJ, Kumagai H, Tajima S, Suganuma N, Suzuki A, Aoki T, et al. Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res 2004; 11:263-74; PMID:15500251; http://dx.doi.org/10.1093/dnares/11.4.263
- Maunoury N, Redondo-Nieto M, Bourcy M, Van de Velde W, Alunni B, Laporte P, Durand P, Agier N, Marisa L, Vaubert D, et al. Differentiation of symbiotic cells and endosymbionts in Medicago truncatula nodulation are coupled to two transcriptome-switches. PLoS One 2010; 5:e9519; PMID:20209049; http://dx.doi.org/10.1371/journal.pone.0009519
- Breakspear A, Liu C, Roy S, Stacey N, Rogers C, Trick M, Morieri G, Mysore KS, Wen J, Oldroyd GE, et al. The root hair “Infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 2014; 26:4680-701; PMID:25527707; http://dx.doi.org/10.1105/tpc.114.133496
- Moreau S, Verdenaud M, Ott T, Letort S, de Billy F, Niebel A, Gouzy J, de Carvalho-Niebel F, Gamas P. Transcription reprogramming during root nodule development in Medicago truncatula. PLoS One 2011; 6:e16463; PMID:21304580; http://dx.doi.org/10.1371/journal.pone.0016463
- Dalla Via V, Narduzzi C, Aguilar OM, Zanetti ME, Blanco FA. Changes in the common bean (Phaseolus vulgaris) transcriptome in response to secreted and surface signal molecules of Rhizobium etli. Plant Physiol 2015; 169:1356-70; PMID:26282238; http://dx.doi.org/10.1104/pp.15.00508
- Jones KM, Sharopova N, Lohar DP, Zhang JQ, VandenBosch KA, Walker GC. Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide-deficient mutant. Proc Natl Acad Sci USA 2008; 105:704-9; PMID:18184805; http://dx.doi.org/10.1073/pnas.0709338105
- D'Antuono AL, Ott T, Krusell L, Voroshilova V, Ugalde RA, Udvardi M, Lepek VC. Defects in rhizobial cyclic glucan and lipopolysaccharide synthesis alter legume gene expression during nodule development. Mol Plant Microbe Interact 2007; 21:50-60; PMID:18052882; http://dx.doi.org/10.1094/MPMI-21-1-0050
