ABSTRACT
Jasmonates (JAs) are a class of plant hormones, essential in plant development and defense. JA induces the interaction of the JA receptor Coronatine Insensitive 1 with jasmonate ZIM-domain (JAZ) proteins, and promotes subsequent JAZs degradation, leading to the release of downstream factors and activation of diverse plant development and defense processes. We recently revealed that the IIIe bHLH transcription factors MYC2, MYC3, MYC4 and MYC5 interact with the MYB transcription factors MYB21 and MYB24 to form the bHLH-MYB complex, and JAZs repress the bHLH-MYB complex to regulate JA-mediated stamen development. Here, we further discuss the different properties of the components of the bHLH-MYB complex in expression pattern and stamen regulation.
The phytohormone jasmonates (JAs) are oxylipin signaling molecules, participating in the control of various aspects of the plant life, such as plant fertility,Citation1,2,3 anthocyanin accumulation,Citation4,5 root growth,Citation6,7,8 fruit ripeningCitation9 and leaf senescence.Citation10,11 They also act as defense signals to mediate plant responses against abiotic and biotic stresses.Citation12,13,14,15 Upon exogenous or endogenous JA induction, the JA receptor protein Coronatine Insensitive 1 interacts with and triggers the degradation of the jasmonate ZIM-domain (JAZ) proteins, which interact with and repress diverse downstream transcription factors to inhibit multiple aspects of JA responses.Citation16-22
The R2R3-MYB transcription factors MYB21 and MYB24 are essential for floral organ development, including petal expansion, stamen development and gynoecium growth, and secondary metabolism in flower.Citation23-26 We previously showed that JAZ proteins interact with MYB21 and MYB24 to regulate JA-mediated stamen development, including filament elongation, anther dehiscence and pollen maturation.Citation24 Recently we reported that the IIIe bHLH transcription factors MYC2, MYC3, MYC4 and MYC5, which are also targets of JAZs,Citation6,7,8,27 function redundantly in promoting stamen development and plant fertility, and regulate JA-mediated stamen development and fertility by forming the bHLH-MYB transcription complex with MYB21 and MYB24.Citation27 Here, we focus on investigating the variant characters of the MYB and bHLH factors in the bHLH-MYB complex.
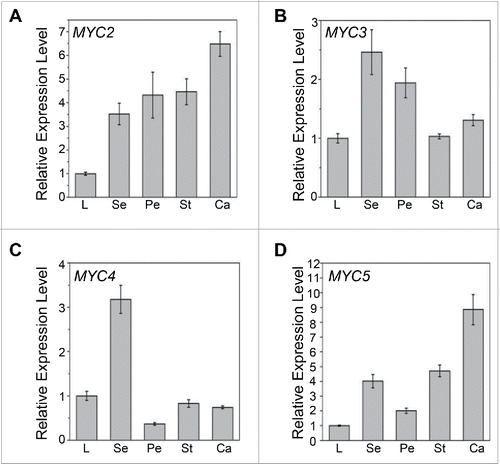
Microarray expression data and histochemical staining analysis have shown that the MYB components (MYB21 and MYB24) of the bHLH-MYB complex are expressed in sepals, petals, stamens and carpels,Citation26,28 supporting their roles in the regulation of floral organ development and secondary metabolism (e.g. sesquiterpene synthesis).Citation23-26 Here, we performed quantitative real-time PCR assays to analyze the expression of the bHLH components of the bHLH-MYB complex in the sepal, petal, stamen and carpel of flower. Expressions of MYC2, MYC3, MYC4 and MYC5 were all detected in stamens (), supporting their role in stamen development and plant fertility.Citation25,26,27 Furthermore, we found that MYC2 and MYC3 exhibited high expression level in all four floral organs (), while MYC4 and MYC5 were preferentially expressed in sepals or carpels respectively (). The expressions of the MYC genes in floral organs indicate that they might also control the development of other flower organs and floral secondary metabolism, which remain to be elucidated. MYC2 has been shown to regulate sesquiterpene synthesis in flower.Citation29
Figure 1. Expression Levels of MYC2, MYC3, MYC4 and MYC5 in Floral Organs. (A-D) Quantitative real-time PCR analyses of MYC2 (A), MYC3 (B), MYC4 (C), and MYC5 (D) in leaves and floral organs of 8-week-old Col-0 wild type using ACTIN8 as the internal control. L, leaf; Se, sepal; Pe, petal; St, stamen; Ca, carpel. Each value is the mean (±SE) of 3 biological replicates.
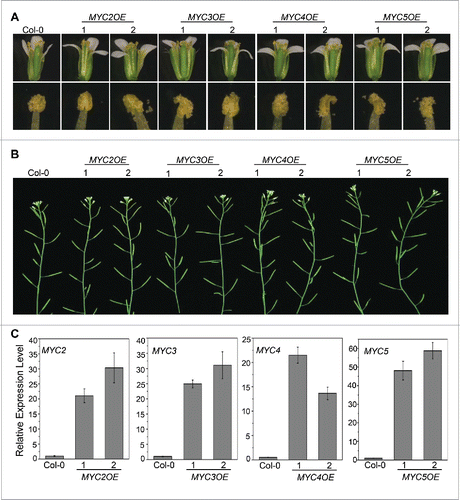
Mutation, inhibition and overexpression of MYB21 and MYB24 obviously inhibit stamen development, demonstrating that proper stamen development requires the appropriate expression level of MYB21 and MYB24.Citation24 Our recent study demonstrated that the malfunction of MYC2, MYC3, MYC4, and MYC5 delays stamen development and reduces fertility.Citation27 Now, we generated MYC2, MYC3, MYC4, and MYC5 overexpressing transgenic plants to investigate whether overexpression of these bHLH factors represses stamen development. Among all the transgenic lines of every gene (101, 68, 71, and 87 overexpressing transgenic lines respectively for MYC2, MYC3, MYC4, and MYC5), none of them showed altered stamen development or plant fertility (2 representative overexpression lines for each gene are shown in ), suggesting that overexpression of MYC2, MYC3, MYC4, or MYC5 does not affect stamen development, which is distinct from that of MYB21 and MYB24.
Figure 2. Overexpression of MYC2, MYC3, MYC4 and MYC5 Cannot Affect Plant Fertility. (A) Comparison of flowers (top) and anther dehiscence (bottom) at floral stage 13 from Col-0 wild type, two transgenic lines respectively for MYC2 (MYC2OE1 and MYC2OE2), MYC3 (MYC3OE1 and MYC3OE2), MYC4 (MYC4OE1 and MYC4OE2), and MYC5 (MYC5OE1 and MYC5OE2). (B) The main inflorescences of the indicated genotypes. (C) Quantitative real-time PCR analyses of MYC2, MYC3, MYC4 or MYC5 in 3-week-old plants of the indicated genotypes using ACTIN8 as the internal control. Each value is the mean (±SE) of three biological replicates.
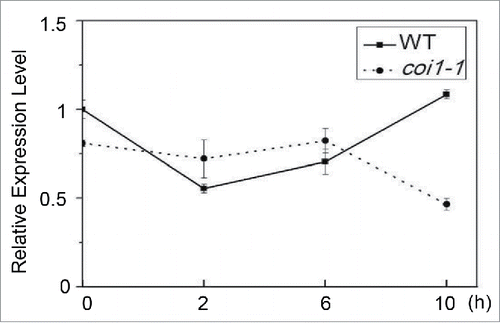
JA regulates the expression of the bHLH and MYB components of the bHLH-MYB complex.Citation7,25,26 The transcript levels of MYB21, MYB24, and MYC2 increased rapidly and significantly to about 20-, 30-, and 16-fold in response to JA treatment,Citation25 whereas the expression of MYC3 and MYC4 were only mildly (˜2 fold) induced by JA.Citation7 Here, we found that the expression of MYC5 cannot be elevated by JA in wild type (), suggesting MYC5 and the other components of the bHLH-MYB complex exhibit different JA-induced expression patterns.
Figure 3. JA Treatment Cannot Induce MYC5 Expression in Arabidopsis. Quantitative real-time PCR analysis of MYC5 expression level using ACTIN8 as the internal control. After JA treatment, MYC5 expression was not significantly raised in WT compared to that in coi1-1, a JA receptor mutant. Each value is the mean (±SE) of three biological replicates.
The MYB factors (MYB21 and MYB24) and bHLH factors (MYC2, MYC3, MYC4 and MYC5) form the bHLH-MYB complex to control JA-mediated stamen development.Citation24,27 Here we noticed that studies have revealed differences existing among these factors: 1) MYC2 and MYC3 exhibit high expression levels in all four floral organs, while MYC4 and MYC5 preferentially express in sepals or carpels, respectively; 2) Overexpression of MYB factors inhibits stamen development, while overexpression of these bHLH factors cannot influence stamen development; 3) The expression of MYB21, MYB24, MYC2, MYC3 and MYC4 can be either significantly or mildly induced by JA, whereas MYC5 cannot. So far, MYB21 and MYB24 have been reported to mainly function in the flower, such as in floral organ development and floral sesquiterpene biosynthesis, while MYC2, MYC3, and MYC4 regulate diverse plant responses, including root growth, leaf senescence, secondary metabolic processes, and plant defense responses, as well as stamen development. As these MYB and bHLH factors share common functions and exhibit differences at the same time, it will be interesting to investigate what other plant responses the bHLH-MYB complex regulates besides stamen development, and what other plant responses the bHLH and MYB factors separately control, and how.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was financially supported by the National Natural Science Foundation of China (Grant 31400253).
References
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K. The Defective in anther dehiscience gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 2001; 13:2191-209; PMID:11595796; http://dx.doi.org/10.1105/tpc.13.10.2191
- Park JH, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 2002; 31:1-12; PMID:12100478; http://dx.doi.org/10.1046/j.1365-313X.2002.01328.x
- Stintzi A, Browse J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 2000; 97:10625-10630; PMID:10973494; http://dx.doi.org/10.1073/pnas.190264497
- Shan X, Zhang Y, Peng W, Wang Z, Xie D. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J Exp Bot 2009; 60:3849-60; PMID:19596700; http://dx.doi.org/10.1093/jxb/erp223
- Yan Y, Christensen S, Isakeit T, Engelberth J, Meeley R, Hayward A, Emery RJ, Kolomiets MV. Disruption of OPR7 and OPR8 reveals the versatile functions of jasmonic acid in maize development and defense. Plant Cell 2012; 24:1420-36; PMID:22523204; http://dx.doi.org/10.1105/tpc.111.094151
- Cheng Z, Sun L, Qi T, Zhang B, Peng W, Liu Y, Xie D. The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-Domain proteins to mediate jasmonate response in Arabidopsis. Mol. Plant 2011; 4:279-88; PMID: 21242320; http://dx.doi.org/10.1093/mp/ssq073
- Fernandez-Calvo P, Chini A, Fernandez-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011; 23:701-15; PMID:21335373; http://dx.doi.org/10.1105/tpc.110.080788
- Niu Y, Figueroa P, Browse J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J Exp Bot 2011; 62:2143-54; PMID:21321051; http://dx.doi.org/10.1093/jxb/erq408
- Kondo S, Tomiyama A, Seto H. Changes of endogenous jasmonic acid and methyl jasmonate in apples and sweet cherries during fruit development. J Am Soc Hortic Sci 2000; 125:282-7
- Reinbothe C, Springer A, Samol I, Reinbothe S. Plant oxylipins: role of jasmonic acid during programmed cell death, defence and leaf senescence. Febs J 2009; 276:4666-81; PMID:19663906; http://dx.doi.org/10.1111/j.1742-4658.2009.07193.x
- Ueda J, Kato J. Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.). Plant Physiol 1980; 66:246-9; PMID:16661414; http://dx.doi.org/10.1104/pp.66.2.246
- Hu P, Zhou W, Cheng Z, Fan M, Wang L, Xie D. JAV1 controls jasmonate-regulated plant defense. Mol Cell 2013; 50:504-15; PMID:23706819; http://dx.doi.org/10.1016/j.molcel.2013.04.027
- Seo JS, Joo J, Kim MJ, Kim YK, Nahm BH, Song SI, Cheong JJ, Lee JS, Kim JK, Choi YD. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J 2011; 65:907-21; PMID:21332845; http://dx.doi.org/10.1111/j.1365-313X.2010.04477.x
- Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci U S A 1998; 95:15107-11; PMID:9844023; http://dx.doi.org/10.1073/pnas.95.25.15107
- Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Close TJ. Large-scale expression profiling and physiological characterization of jasmonic acid-mediated adaptation of barley to salinity stress. Plant Cell Environ 2007; 30:410-21; PMID:17324228; http://dx.doi.org/10.1111/j.1365-3040.2006.01628.x
- Abuqamar S, Chai MF, Luo H, Song F, Mengiste T. Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell 2008; 20:1964-83; PMID:18599583; http://dx.doi.org/10.1105/tpc.108.059477
- Chico JM, Chini A, Fonseca S, Solano R. JAZ repressors set the rhythm in jasmonate signaling. Curr Opin Plant Biol 2008; 11:486-94; PMID:18653378; http://dx.doi.org/10.1016/j.pbi.2008.06.003
- Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007; 448:666-71; PMID:17637675; http://dx.doi.org/10.1038/nature06006
- Hu Y, Jiang L, Wang F, Yu D. Jasmonate regulates the inducer of CBF expression-c-repeat binding factor/dre binding factor1 cascade and freezing tolerance in arabidopsis. Plant Cell 2013; 25:2907-24; PMID:23933884; http://dx.doi.org/10.1105/tpc.113.112631
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D. The Jasmonate-ZIM-Domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 2011; 23:1795-814; PMID:21551388; http://dx.doi.org/10.1105/tpc.111.083261
- Song S, Qi T, Fan M, Zhang X, Gao H, Huang H, Wu D, Guo H, Xie D. The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet 2013; 9:e1003653; PMID:23935516; http://dx.doi.org/10.1371/journal.pgen.1003653
- Yan Y, Stolz S, Chetelat A, Reymond P, Pagni M, Dubugnon L, Farmer EE. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 2007; 19:2470-83; PMID:17675405; http://dx.doi.org/10.1105/tpc.107.050708
- Reeves PH, Ellis CM, Ploense SE, Wu MF, Yadav V, Tholl D, Chételat A, Haupt I, Kennerley BJ, Hodgens C, et al. A Regulatory Network for Coordinated Flower Maturation. PLoS Genet 2012; 8(2):e1002506; PMID:22346763; http://dx.doi.org/10.1371/journal.pgen.1002506
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 2011; 23:1000-13; PMID:21447791; http://dx.doi.org/10.1105/tpc.111.083089
- Mandaokar A, Thines B, Shin B, Lange BM, Choi G, Koo YJ, Yoo YJ, Choi YD, Choi G, Browse J. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J 2006; 46:984-1008; PMID:16805732; http://dx.doi.org/10.1111/j.1365-313X.2006.02756.x
- Cheng H, Song S, Xiao L, Soo H, Cheng Z, Xie D, Peng J. Gibberellin Acts through Jasmonate to Control the Expression of MYB21, MYB24, and MYB57 to Promote Stamen Filament Growth in Arabidopsis. Plos Genet 2009; 5(3):e1000440; PMID:19325888; http://dx.doi.org/10.1371/journal.pgen.1000440
- Qi T, Huang H, Song S, Xie D. Regulation of Jasmonate-Mediated Stamen Development and Seed Production by a bHLH-MYB Complex in Arabidopsis. Plant Cell 2015; 27:1620-33; PMID:26002869; http://dx.doi.org/10.1105/tpc.15.00116
- Reeves PH, Ellis CM, Ploense SE, Wu MF, Yadav V, Tholl D, Chetelat A, Haupt I, Kennerley BJ, Hodgens C, et al. A regulatory network for coordinated flower maturation. PLoS Genet 2012; 8:e1002506; PMID:22346763; http://dx.doi.org/10.1371/journal.pgen.1002506
- Hong G, Xue X, Mao Y, Wang L, Chen X. Arabidopsis MYC2 Interacts with DELLA Proteins in Regulating Sesquiterpene Synthase Gene Expression. Plant Cell 2012; 24:2635-48; PMID:22669881; http://dx.doi.org/10.1105/tpc.112.098749
