Abstract
We identified virulence-related effectors of a hemibiotrophic fungal pathogen Colletotrichum orbiculare, and found that a novel interface was generated by a biotrophic interaction between C. orbiculare and the host cucumber, in which the effectors secreted from the pathogen accumulated preferentially. The interface was located around the biotrophic primary hyphal neck. Here, we showed that C. orbiculare also developed this interface in a biotrophic interaction with melon, which belongs to Cucurbitaceae. Furthermore, C. orbiculare developed interface in the interaction with a susceptible plant, Nicotiana benthamiana, which is distantly related to Cucurbitaceae, suggesting that the spatial regulation strategy for effectors in C. orbiculare is not specific to cucumber; rather, it is conserved among the various plants that are susceptible to this pathogen.
Fungal phytopathogens cause severe damage to food and cash crop productions worldwide.Citation1 Hemibiotrophic fungal pathogens establish biotrophic interactions with host plants, followed by a destructive necrotrophy. In this process, they synthesize and secrete an arsenal of virulence proteins called “effectors” to manipulate their host environmental status; in particular, biotrophs and hemibiotrophs utilize effectors to keep their host alive.Citation2–5
Characteristic fungus–plant interfaces that accumulate effectors in biotrophy were previously reported in hemibiotrophs, the rice blast fungus Magnaporthe oryzaeCitation6,7 and the crucifer anthracnose fungus Colletotrichum higginsianum,Citation8 as well as in obligate biotrophs.Citation9,10 Interestingly, there are a wide variety of differences in shape and location among these interfaces, suggesting the presence of diversity in the effector secretion strategy of the pathogens for successful infection of their corresponding host plants, which is probably generated via each pathogen–plant coevolution.
Colletotrichum orbiculare, a cucumber anthracnose fungus, develops melanized appressoria for penetration and exhibits a hemibiotrophic lifestyle at postinvasive phases.Citation11,12 Analyses of the genome sequence of C. orbiculare uncovered the presence of numerous effector candidate genes in this pathogen.Citation13 Recently, we identified multiple virulence effectors of C. orbiculare, designated NIS1, CoDN3 (hereafter called DN3) and CoMC69 (hereafter called MC69), that are expressed in biotrophic hyphae of the pathogen.Citation14,15 NIS1 was found to induce cell death in Nicotiana benthamiana via a functional screening based on Agrobacterium-mediated transient expression, whereas DN3 was identified as an effector that suppresses NIS1-induced cell death.Citation14 MC69 was shown to be essential for the full virulence of C. orbiculare.Citation15
To identify possible effector-related interfaces generated via the interaction between C. orbiculare and the host, cucumber, we examined the spatial and temporal dynamics of the effectors identified during infection of the host plant, cucumber, by C. orbiculare.Citation16 Live-cell imaging of fluorescence-tagged effectors revealed the presence of a strong ring-like signal of the tagged effectors in the biotrophic interaction between C. orbiculare and cucumber.Citation16 The visualization of the fungal cytoplasm or cell-wall components together with the effectors indicated that the ring signal was not present inside C. orbiculare; rather, it was located around the biotrophic primary hyphal neck, indicating that the ring signal of the effector is present in the interfacial region between cucumber and C. orbiculare.Citation16 This result was further supported by transmission electron microscopy (TEM) analyses that were focused on the neck region of biotrophic primary hyphae.Citation16 We did not detect the effector-related ring signal around the pseudobiotrophic hyphal neck of C. orbiculare that was formed in an artificial substratum, which strongly supports the requirement of a fungus–host plant interaction for the development of the interface. Moreover, a photobleaching analysis of the ring signal of fluorescence-tagged effector suggested that effector proteins are continuously synthesized and delivered to the interface in biotrophy.Citation16 These findings revealed that C. orbiculare actively secretes the effectors toward the novel plant–pathogen interfacial region located around the neck of the primary biotrophic hyphae that developed in host cucumber cells ().
Figure 1. C. orbiculare secretes effectors at the interfacial region, as visualized by DN3:mCherry as a ring signal, during the susceptible interaction with multiple plant species. (A) Schematic model of the spatial secretion strategy of the effectors in C. orbiculare for plant infection. Using melanized appressorium, C. orbiculare penetrates the cell wall (CW) of susceptible plants and subsequently develops biotrophic invasive hyphae. The developed biotrophic invasive hyphae are surrounded by the extrainvasive hyphal membrane (EIHM), which is continuous with the plant plasma membrane (PM). C. orbiculare preferentially secretes virulence-related effectors, such as DN3, at the interfacial region located around the neck region of primary biotrophic invasive hyphae, which is called ring-signal interfacial region. C. orbiculare also likely secretes effectors at randomly distributed interfacial bodies. (B) Inoculation of the C. orbiculare wild-type (WT) strain 104-T on cotyledons of both melon and cucumber. A conidial suspension (5 × 105 conidia/mL) of 104-T was drop-inoculated on cotyledons of both melon and cucumber, and the inoculated plants were incubated for 7 d. (C) Focal accumulation of the DN3 effector, which is secreted by C. orbiculare during the susceptible interaction with multiple plant species. A C. orbiculare WT strain expressing the DN3 effector:mCherry fusion protein under the TEF promoter was inoculated on melon cotyledons or N. benthamiana leaves. Images were taken at 3 d postinoculation (dpi) for melon and at 4 dpi for N. benthamiana using confocal laser scanning microscopy. Bars = 10 μm.
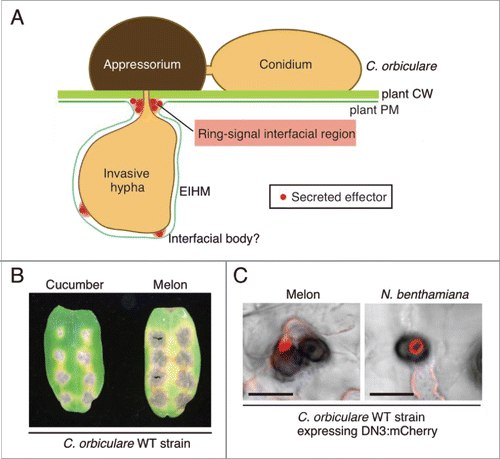
In the biotrophic interaction between the crucifer anthracnose fungus C. higginsianum and Arabidopsis thaliana, effector-related interfacial regions, which are named interfacial bodies, were detected between biotrophic hyphae of C. higginsianum and Arabidopsis plasma membranes by fluorescent labeling of effectors and immuno-TEM analyses.Citation8 Consistently, we also observed the accumulation of effectors as multiple punctate structures located around the biotrophic hyphal surface of C. orbiculare (),Citation16 which implies that C. orbiculare also delivers the effectors to putative interfaces on the hyphal surface, in addition to the ring-signal interface at postinvasive stages. When appressoria did not develop biotrophic hyphae, we frequently detected single-dot signals of effectors at the bottom of appressoria.Citation16 In the C. higginsianum–Arabidopsis interaction, effector accumulation in appressorial penetration pores before host invasion has been found as an interface at the preinvasion stage.Citation8 These observations suggest that the focal secretion of effectors in the appressorial penetration pore is likely a common strategy used by Colletotrichum species for successful penetration and subsequent development of invasive hyphae. The entry trail of nonadapted fungi is mostly terminated by preinvasive immune responses of nonhost plants.Citation17,18 Effector delivery at the appressorial pore of Colletotrichum fungi might play a critical role in the suppression of preinvasion defenses against nonadapted pathogens.
Although we revealed that C. orbiculare secretes the effectors at the unique interface developed between C. orbiculare and cucumber, it is unclear whether this phenomenon is specific to the biotrophic interaction between C. orbiculare and cucumber, or whether it common among the interactions between C. orbiculare and other susceptible plants. To assess this point, we first investigated the interaction between C. orbiculare and melon, which belongs to Cucurbitaceae, similar to cucumber. To assess whether C. orbiculare infects melon effectively, we drop-inoculated a conidial suspension of the C. orbiculare strain 104-T (MAFF240422) on cotyledons of melon plants (Cucumis melo cv Lennon). As a control, we also inoculated a conidial suspension on cucumber cotyledons (Cucumis sativus L. cv Suyo). We found that the melon cotyledons were more susceptible to the C. orbiculare strain 104-T compared with the cucumber cotyledons (). Subsequently, we inoculated a C. orbiculare transgenic strain expressing mCherry-tagged DN3 on the lower surface of melon cotyledons. The strain constitutively expresses the tagged DN3 effector under the TEF promoter of Aureobasidium pullulans.Citation19 At 3 d postinoculation (dpi), we peeled off the epidermal layers of melon cotyledons, and the peeled layers were subjected to confocal microscopic analysis. We found the typical ring signal of the tagged effectors around the neck of biotrophic primary hyphae (). In contrast, it was difficult to see the typical ring signal of the tagged effectors at 4 dpi of melon cotyledons, whereas ring signal was typically observed at 4 dpi of cucumber cotyledons.Citation16 We found that C. orbiculare enters the necrotrophic phase as early as 4 dpi of melon, which is likely consistent with the finding that melon cotyledons were more susceptible to C. orbiculare than were cucumber cotyledons. This finding reveals that C. orbiculare commonly develops the ring-signal-related interface in susceptible interactions with multiple Cucurbitaceous plants. It also indicates that the development of the interface is tightly related to the biotrophic phase, even when the timing of the biotrophic phase is changed.
It is also known that C. orbiculare strains, including 104-T, are able to infect N. benthamiana plants.Citation20,21 N. benthamiana belongs to the Solanaceae family, which is distantly related to Cucurbitaceous plants. We asked whether C. orbiculare develops the ring-signal interface in N. benthamiana when the pathogen establishes the biotrophic phase in this particular plant species. We inoculated the C. orbiculare strains expressing mCherry-tagged DN3 on N. benthamiana leaves. Notably, we also detected the ring signal of the tagged effectors in the interaction sites of C. orbiculare with N. benthamiana at 3–4 dpi (). Thus, the spatial regulation strategy of the effectors in C. orbiculare is not limited to Cucurbitaceous plants; rather, it is conserved among the broad range of plants that are susceptible to C. orbiculare. Thus, the data presented here suggest that C. orbiculare probably secretes an unidentified molecule that triggers the development of the ring-signal interface in its susceptible plants, which are not restricted to Cucurbitaceous plants, but extends to N. benthamiana.
What is the physiological role of the ring-signal-related interfacial region around the neck of the primary invasion hyphae of C. orbiculare that is detected during the infection of multiple susceptible plants? The ring-signal interface develops de novo during the formation of biotrophic hyphae. Furthermore, effector delivery to the interface was tightly coupled to early biotrophic expression of effector genes,Citation16 which implies biotrophy-associated functions, such as the maintenance of biotrophy and/or suppression of postinvasion defenses until the switch to necrotrophy. Consistent with this idea, it was difficult to detect the ring signal at 4 dpi of melon cotyledons when the pathogen had already entered the necrotrophic phase, which further supports the importance of the ring-signal-related interface for biotrophy in the C. orbiculare species.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported in part by Grants-in-Aid for Scientific Research (15H04457 and 15H05780) from the Ministry of Education; Culture, Sports, Science, and Technology (MEXT) of Japan; by the Council for Science, Technology and Innovation (CSTI), Cross-ministerial Strategic Innovation Promotion Program (SIP), “Technologies for creating next-generation agriculture, forestry and fisheries” (Funding agency: Bio-oriented Technology Research Advancement Institution, NARO), by the Science and Technology Research Promotion Program for the Agriculture, Forestry, Fisheries, and Food industry; and by a grant-in-aid from the Institution for Fermentation, Osaka.
References
- Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012; 484:186-194; PMID:22498624; http://dx.doi.org/10.1038/nature10947
- Kamoun S. Groovy times: Filamentous pathogen effectors revealed. Curr Opin Plant Biol 2007; 10:358-65; PMID:17611143; http://dx.doi.org/10.1016/j.pbi.2007.04.017
- Koeck M, Hardham AR, Dodds PN. The role of effectors of biotrophic and hemibiotrophic fungi in infection. Cell Microbiol 2011; 13:1849-57; PMID:21848815; http://dx.doi.org/10.1111/j.1462-5822.2011.01665.x
- Dou D, Zhou JM. Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 2012; 12:484-95; PMID:23084917; http://dx.doi.org/10.1016/j.chom.2012.09.003
- Rafiqi M, Ellis JG, Ludowici VA, Hardham AR, Dodds PN. Challenges and progress towards understanding the role of effectors in plant-fungal interactions. Curr Opin Plant Biol 2012; 15:477-82; PMID:22658704; http://dx.doi.org/10.1016/j.pbi.2012.05.003
- Mosquera G, Giraldo MC, Khang CH, Coughlan S, Valent B. Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as biotrophy-associated secreted proteins in rice blast disease. Plant Cell 2009; 21:1273-90; PMID:19357089; http://dx.doi.org/10.1105/tpc.107.055228
- Khang CH, Berruyer R, Giraldo MC, Kankanala P, Park SY, Czymmek K, Kang S, Valent B. Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 2010; 22:1388-1403; PMID:20435900; http://dx.doi.org/10.1105/tpc.109.069666
- Kleemann J, Rincon-Rivera LJ, Takahara H, Neumann U, van Themaat EV, van der Does HC, Hacquard S, Stüber K, Will I, Schmalenbach W, et al. Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum. PLoS Pathog 2012; 8:e1002643; PMID:22496661; http://dx.doi.org/10.1371/journal.ppat.1002643
- Catanzariti AM, Dodds PN, Lawrence GJ, Ayliffe MA, Ellis JG. Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 2006; 18:243-56; PMID:16326930; http://dx.doi.org/10.1105/tpc.105.035980
- Micali CO, Neumann U, Grunewald D, Panstruga R, O'Connell R. Biogenesis of a specialized plant-fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cell Microbiol 2011; 13:210-226; PMID:20880355; http://dx.doi.org/10.1111/j.1462-5822.2010.01530.x
- Perfect SE, Hughes HB, O'Connell RJ, Green JR. Colletotrichum: A model genus for studies on pathology and fungal-plant interactions. Fungal Genet Biol 1999; 27:186-198; PMID:10441444; http://dx.doi.org/10.1006/fgbi.1999.1143
- Kubo Y, Takano Y. Dynamics of infection-related morphogenesis and pathogenesis in Colletotrichum orbiculare. J Gen Plant Pathol 2013; 79:233-242; http://dx.doi.org/10.1007/s10327-013-0451-9
- Gan P, Ikeda K, Irieda H, Narusaka M, O'Connell RJ, Narusaka Y, Takano Y, Kubo Y, Shirasu K. Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. New Phytologist 2013; 197:1236-49; PMID:23252678; http://dx.doi.org/10.1111/nph.12085
- Yoshino K, Irieda H, Sugimoto F, Yoshioka H, Okuno T, Takano Y. Cell death of Nicotiana benthamiana is induced by secreted protein NIS1 of Colletotrichum orbiculare and is suppressed by a homologue of CgDN3. Mol Plant-Microbe Interact 2012; 25:625-36; PMID:22352720; http://dx.doi.org/10.1094/MPMI-12-11-0316
- Saitoh H, Fujisawa S, Mitsuoka C, Ito A, Hirabuchi A, Ikeda K, Irieda H, Yoshino K, Yoshida K, Matsumura H, et al. Large-scale gene disruption in Magnaporthe oryzae identifies MC69, a secreted protein required for infection by monocot and dicot fungal pathogens. PLoS Pathog 2012; 8:e1002711; PMID:22589729; http://dx.doi.org/10.1371/journal.ppat.1002711
- Irieda H, Maeda H, Akiyama K, Hagiwara A, Saitoh H, Uemura A, Terauchi R, Takano Y. Colletotrichum orbiculare secretes virulence effectors to a biotrophic interface at the primary hyphal neck via exocytosis coupled with SEC22-mediated traffic. Plant Cell 2014; 26:2265-81; PMID:24850852; http://dx.doi.org/10.1105/tpc.113.120600
- Lipka U, Fuchs R, Lipka V. Arabidopsis nonhost resistance to powdery mildews. Curr Opin Plant Biol 2008; 11:404-11; PMID:18499508; http://dx.doi.org/10.1016/j.pbi.2008.04.004
- Shimada C, Lipka V, O'Connell R, Okuno T, Schulze-Lefert P, Takano Y. Nonhost resistance in Arabidopsis-Colletotrichum interactions acts at the cell periphery and requires actin filament function. Mol Plant Microbe Interact 2006; 19:270-279; PMID:16570657; http://dx.doi.org/10.1094/MPMI-19-0270
- Vanden Wymelenberg AJ, Cullen D, Spear RN, Schoenike B, Andrews JH. Expression of green fluorescent protein in Aureobasidium pullulans and quantification of the fungus on leaf surfaces. Biotechniques 1997; 23:686-690; PMID:9343693.
- Shen S, Goodwin PH, Hsiang T. Infection of Nicotiana species by the anthracnose fungus, Colletotrichum orbiculare. Eur J Plant Pathol 2001;107:767-73;http://dx.doi.org/10.1023/A:1012280102161
- Takano Y, Takayanagi N, Hori H, Ikeuchi Y, Suzuki T, Kimura A, Okuno T. A gene involved in modifying transfer RNA is required for fungal pathogenicity and stress tolerance of Colletotrichum lagenarium. Mol Microbiol 2006; 60:81-92; PMID:16556222; http://dx.doi.org/10.1111/j.1365-2958.2006.05080.x
