ABSTRACT
Phytoplasmas are among the most recently discovered plant pathogenic microorganisms so, many traits of the interactions with host plants and insect vectors are still unclear and need to be investigated. At now, it is impossible to determine the precise sequences leading to the onset of the relationship with the plant host cell. It is still unclear how phytoplasmas, located in the phloem sieve elements, exploit host cell to draw nutrition for their metabolism, growth and multiplication. In this work, basing on microscopical observations, we give insight about the structural interactions established by phytoplasmas and the sieve element plasma membrane, cytoskeleton, sieve endoplasmic reticulum, speculating about a possible functional role.
Phytoplasmas, phloem-residing biotrophic prokaryotes that belong to the Mollicutes, are among the latest discovered plant-pathogenic microorganisms.Citation1 They initially received modest attention so that many major traits of their interaction with host plants and insect vectors still require thorough investigation. Due to difficulties in cultivating phytoplasmas in vitro,Citation2 however, it has turned out to be impossible to date to determine the precise sequence and nature of the events that establish the relationship with the host plant.
Recently,Citation3 we described an extensive ultrastructural re-organization of sieve elements (SEs) in tomato plants (Solanum lycopersicum L.) affected by stolbur, the disease associated with the ‘Candidatus Phytoplasma solani’ (‘Ca. P. solani’). We showed the presence of membranous connections between phytoplasma and SE plasma membrane, and a close association of phytoplasma cells with the sieve-element reticulum (SER) and SE cytoskeleton.Citation3 Based on further electron-microscopic observations using conventionally prepared sections (see Citation3), we speculate here on the order of events in phytoplasma colonization and on the functional significance of the structural interaction between phytoplasmas and the SE components mentioned before.
Phytoplasma inoculation and initial floating in the SE lumen
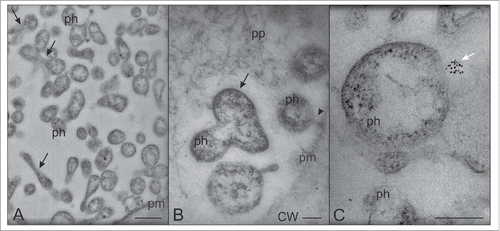
After direct inoculation inside the SEs by the stylects of insect vectors,Citation4 phytoplasmas probably float freely in the SE lumen having mostly round, sometimes elongated shapes (). Several of the phytoplasmas in the lumen exhibit actin labeling, which discloses unipolar actin fields ( and Citation3). The presence of host-cell actin filaments on the phytoplasma membrane surface may enable the phytoplasmas to adhere to SE-structures and then anchor to the SE plasma membraneCitation3 in analogy to events described for other pathogen/host cell interactions.Citation5,6 We also observed free-floating dividing phytoplasmas in the lumen (), of which it is unclear if the division occurs before or after phytoplasma anchoring.
Figure 1. Transmission electron microscopy (TEM) micrographs of phytoplasmas floating in the SE lumen. (A,B) Phytoplasmas are mostly roundish, sometimes elongated; a few are dividing (black arrows). (C) Aggregates of SE actin form unipolar fields on the phytoplasma surface in the SE lumen (white arrow). The arrowhead in (B) indicates the attachment of a phytoplasma to the SE plasma membrane. In (A), the bar corresponds to 500 nm; in (B) and (C) the bars correspond to 200 nm. CW: cell wall; ph: phytoplasma; pm: plasma membrane; pp phoem protein.
Phytoplasma adhesion to host cell structures and anchoring to the SE plasma membrane
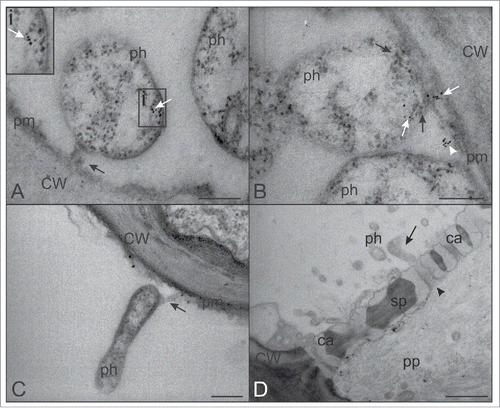
Bacterial adhesion to host cell structures is a fundamental step in the pathogenic process. Subsequent to inoculation by their vectors phytoplasmas supposedly initiate the infection cycle by anchoring to the host cell plasma membrane.Citation7 The discovery of genes encoding putative membrane proteins carrying an adhesion motif on the surface of phytoplasmasCitation8-12 is supportive of phytoplasma adhesion to SE substructures. We highlight here that, in most cases, the “adhesion structure” that anchors the phytoplasma body to the SE plasma membrane is tubular, 30–40 nm in width and orientated perpendicularly to SE plasma membrane (). The central part appears mostly electron-translucent (). Sometimes, the tubular corridor shows a widened end toward the SE plasma membrane () so that the structure is spatially funnel-like. However, differences in morphology of the “adhesion structure” may depend on the orientation of sectioning.
Figure 2. TEM micrographs showing phytoplasmas and the adjoining SE plasma membrane. (A) A tubular “adhesion structure,” connecting the phytoplasma body to the SE plasma membrane is visible (black arrow). SE actin (white arrow in inset i, upper left) is aggregated on phytoplasma cell membrane at the opposite side of the “adhesion structure” (black arrow). (B) Actin (white arrows) is also located in the proximity of the “adhesion structure” (black arrows) and close to the SE plasma membrane (white arrowhead). (C) Elongate phytoplasmas attached (black arrow) to the plasma membrane. (D) Phytoplasmas near the sieve pores. The elongate shape of the phytoplasma body (black arrow), similar in dimension to the sieve pore (black arrowhead) diameter, is required to move from one SE to the next one. In (A), (B) and (C) the bars correspond to 200 nm; in (D), the bar corresponds to 1μm. Ca: callose; CW: cell wall; ph: phytoplasma; pm: plasma membrane; pp: phloem protein; sp: sieve plate.
Nothing is known regarding the molecular architecture or the functional significance of phytoplasma “adhesion structure.” In Mycoplasmas (Mollicutes phylogenetically related to phytoplasmas), in particular in Mycoplasma pneumoniae, a complex, distinct polar extension of the cell body that mediates both adherence to host cell and gliding motility, has been amply studied.Citation13 Dense clusters of adhesion proteins at the tip of this polar extension (named terminal organelle) have been described.Citation13 A putative membrane protein containing a Mollicutes-adhesion-motif (MAM) was recently identified in onion yellow phytoplasmaCitation12 indicating a potential role in anchoring to the host SE plasma membrane.
In addition to phytoplasma surface proteins, host-cell actin is likely engaged in anchoring of phytoplasmas. Phytoplasmas impose an intense remobilization of host-cell actin.Citation3 This indicates that actin on the phytoplasma surface is probably derived from the actin network that has recently been identified in SEs.Citation14 In support of a prominent role in anchoring, actin is present on the SE plasma membrane in the proximity of phytoplasma “adhesion structure” ().
Actin-mediated phytoplasma movement and spread
Adhesion and anchoring may have similarities with the action of diverse pathogenic bacteria,Citation15 which induce early accumulation of actin cytoskeletal complexes at the plasma membrane of the host cell.Citation16 Hence, actin-anchored corridors may play a role in active displacement of phytoplasmas, in particular through the sieve pores, the cellular bottlenecks between SEs. Using molecular approaches, Galetto et al.Citation17 and Boonrod et al.Citation18 demonstrated that phytoplasmas are able to bind to vector and host-plant actin filaments.
As for the intracellular movements inside the host, it is known that intracellular pathogens have evolved very similar modes of motility, dependent on the unipolar polymerization of host actin.Citation19 Many microbial pathogens have evolved the ability to exploit the host-cell actin cytoskeleton to disseminate within and through infected tissues.Citation15,20 The contact between phytoplasmas and plant host actin has been recently demonstrated in situ.Citation3 Actin is localized at one side of a large portion of the phytoplasma bodies floating in the SE lumen (). Actin is simultaneously present on the phytoplasma membrane and the SE plasma membrane,Citation3 presumably to displace phytoplasmas against the mass flow () and/or to facilitate passage of phytoplasmas through the sieve pores.Citation3 As the phytoplasma diameterCitation4 is appreciably larger than sieve pore diameter,Citation21 phytoplasmas must modify their morphology when moving to the next SE. The elongation required to pass the narrow pores may also be dependent on actin activities (). Thus, the actin-binding capability probably has a multiple function in phytoplasma spread.
Other potential functions of the adhesion structure
It is still unclear, how phytoplasmas withdraw resources from the host for their metabolism, growth and multiplication. It seems obvious that anchoring of pathogen and host cell and the subsequent formation of cytoplasmic corridors () facilitate pathogen nutrition and the exchange of molecular signals responsible for pathogen multiplication and spread through the tissues, to ensure host colonization. Despite the apparent logic, it is hard to conceive how the corridors tethered to the plasma membrane () function in phytoplasma nutrition, since open connections to the SE for bulk flow seem to be absent and substances can be only absorbed from the nutrition-poor SE apoplast.
Phytoplasmas and sieve-element reticulum: nourishment and multiplication?
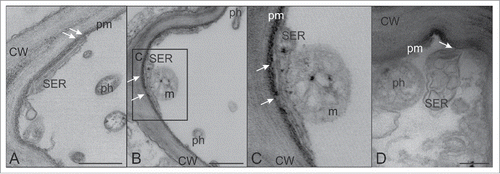
A stable parietal position of the SER, tethered by minute clamps, has been described for SEs in well-preserved sections of Vicia faba and S. lycopersicum.Citation22 In stolbur-infected SEs of tomato, it was not easy to visualize the minute anchors attaching the distorted SER to SE plasma membrane. These minute structures are likely still present in infected SEs (), but in a few cases, SER was observed to be partially detached from SE plasma membrane ().
Figure 3. TEM micrographs showing sieve endoplasmic reticulum (SER) and phytoplasmas. (A,B and inset C). In infected SEs, the SER stays in parietal position by means of minute anchoring structures (arrows). (D) The SER shows swollen, enlarged and distorted cisternae and a partial detachment from the SE plasma membrane (arrow). In (A) and (B) the bars correspond to 500 nm; in (D) the bar corresponds to 200 nm. CW: cell wall; m: mitochondrion; ph: phytoplasma; pm: plasma membrane.
Apart from their contacts with the SER plasma membrane, phytoplasmas also establish relationships with the (distorted) SER,Citation3 which is considered an important source of proteins and metabolites available for intracellular pathogens.Citation23 Like other pathogenic intracellular bacteria,Citation3,24 phytoplasmas induce stress upon the ER followed by the unfolded protein response (UPR).Citation3 The UPR induction could reflect a means through which pathogens gain nutrients from the strongly distorted ER of the host cell.Citation25
All in all, SER-stacks may be more serious candidates for phytoplasma nutrition stores than the SE apoplast. However, open physical contacts were not observed between phytoplasmas and SER stacks; phytoplasmas seem to solely rest close to the contorted SERCitation3 (). Strikingly, actin labeling is always absent on the surface of phytoplasmas adhered to the SERCitation3 (Buxa et al. 2015; ). In conclusion, the proximity of the pathogens and host SER may facilitate the scavenging of nutrients, but the mechanisms of interaction between phytoplasmas and SER are unknown, and demand further in-depth investigation.
It is further imperative to realize that companion cells, providing all compounds needed for sieve-element maintenance via pore-plasmodesmata units,Citation26 must largely contribute to the supply of metabolites and macromolecular building blocks to phytoplasmas.
Concluding Remarks
To acquire host-derived nutrients, obligately intracellular pathogens must exploit the internal stores of host cells by establishing relationships with the host-cell compartments. The distribution of phytoplasmas inside the SE and the connection to the SE substructures allowed us to speculate on the involvement of SE organelles in the pathogen life cycle. The number of phytoplasma bodies found in each SE compartment is reported in . Phytoplasmas were counted and recorded according to their position: attached to the plasma membrane, located in the proximity of SER and floating in the lumen (). Quantitative analyses have been performed on leaves, sampled from fully-symptomatic plants. The majority of phytoplasmas (approx. 75%) occurs in the SE lumen, some (approx. 20%) are attached to the plasma membrane and few (approx. 5%) are in the proximity of the SER. The possible relationships of phytoplasmas with SE organelles are summarized in a scheme (). Even if the events that determine the dynamics of nutrition, proliferation and spread of phytoplasmas within their host are still not understood, bacterial distribution inside the infected SE may give a first glimpse on the interaction of the phytoplasmas with different host-cell compartments in keeping with the pathogen life cycle. The exact order of events is uncertain (), but it seems that part of the phytoplasmas is equipped with a unipolar actin field that enables to adhere to SE membranes. It is unclear if such contacts to the plasma membrane are compulsory. Part of the phytoplasmas may move on to the sieve pore to become elongate, other may release the contact tubule and move to the SER, where they withdraw nutrition from the host (contact/relation with SER) in order to multiply eventually.
Figure 4. (A) Schematic representation of plasma membrane, actin location (red) and SER in non-infected SEs. The plasma membrane is appressed to the SE wall. SER stacks ordered parallel to the plasma membrane, regular in shape, are connected to each other and to plasma membrane by tiny anchors and are accompanied by longish actin filaments (in red: actin, in green: SER proteins). (B) Possible interrelationships of phytoplasmas with SE substructures. The order of “phytoplasma processing” is represented by arrows. The SE plasma membrane, SER stacks and actin filaments are re-arranged following phytoplasma infection. The SE plasma membrane becomes undulated or invaginated, After injection into the SE by insect vectors (a), some of the phytoplasmas locate to the surface of the distorted SER (c,d) that is partially detached from the plasma membrane. Others become equipped with a unipolar SE actin field (b) that enables them to attach to the plasma membrane by “tubular adhesion corridors.” The corridors may be involved in nutrition and signaling and active displacement of phytoplasmas along the plasma membrane (e) and through the sieve pores (f). As indicated by several question marks, the sequence of events is unknown as well as whether the various stages occur in a compulsory order.
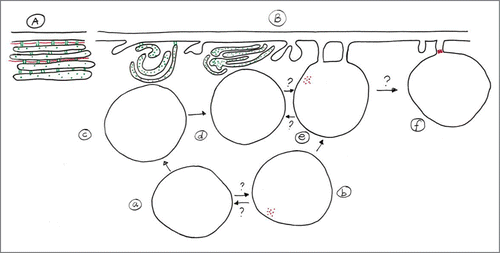
Table 1. Subcellular distribution of phytoplasmas in infected sieve-elements of tomato.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors are grateful to Mrs. Maud Quaedvlieg for drawing .
References
- Doi Y, Teranaka M, Yora K, Asuyama H. Mycoplasma or P.L.T. group-like microorganisms found in the phloem elements of plants infected with mulberry dwarf, potato witches'-broom, aster yellows or paulownia witches'-broom. Ann Phytopathol Soc Japan 1967; 33:259-66.1; http://doi.org/10.3186/jjphytopath.33.259
- Contaldo N, Bertaccini A, Paltrinieri S, Windsor HM, Windsor GD. Axenic culture of plant pathogenic phytoplasmas. Phytopathol Med 2012; 51:607-17; http://dx.doi.org/10.14601/Phytopathol_Mediterr-11773
- Buxa SV, Degola F, Polizzotto R, De Marco F, Loschi A, Kogel KH, di Toppi LS, van Bel AJ, Musetti R. Phytoplasma infection in tomato is associated with re-organization of plasma membrane, ER stacks, and actin filaments in sieve elements. Front Plant Sci 2015; 6:650; PMID:26347766; http://dx.doi.org/10.3389/fpls.2015.00650
- Hogenhout SA, Oshima K, Ammar E, Kakizawa S, Kingdom HN, Namba S. Phytoplasmas: bacteria that manipulate plants and insects. Mol Plant Pathol 2008; 9:403-23; PMID:18705857; http://dx.doi.org/10.1111/j.1364-3703.2008.00472.x
- Giron JA, Lange M, Baseman JB. Adherence, fibronectin binding and induction of cytoskeleton reorganization in cultured human cells by Mycoplasma penetrans. Infect Immun 1996; 64:197-208; PMID:8557340
- Koutsoudis MD, Tsaltas D, Minogue TD, von Bodman SB. Quorum-sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii. Proc Natl Acad Sci USA 2006; 103:5983-8; PMID:16585516; http://dx.doi.org/10.1073/pnas.0509860103
- Marcone C. Movement of phytoplasmas and the development of disease in the plant. In: Phytoplasmas: genomes, plant hosts and vectors, Wallingford, UK: CABI Publishing, 2010; pp 114-131.
- Kube M, Mitrovic J, Duduk B, Rabus R, Seemüller E. Current view on phytoplasma genomes and encoded metabolism. Scie World J 2012; doi:10.1100/2012/185942
- Bai X, Zhang J, Ewing A, Miller SA, Jancso Radek A, Shevchenko DV, Tsukerman K, Walunas T, Lapidus A, Campbell JW, et al. Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J Bacteriol 2006; 188:3682-96; PMID:16672622; http://dx.doi.org/10.1128/JB.188.10.3682-3696.2006
- Neriya Y, Maejima K, Nijo T, Tomomitsu T, Yusa A, Himeno M, Netsu O, Hamamoto H, Oshima K, Namba S. Onion yellow phytoplasma P38 protein plays a role in adhesion to the hosts. FEMS Microbiol Lett 2014; 361:115-22; PMID:25302654; http://dx.doi.org/10.1111/1574-6968.12620
- Oshima K, Kakizawa S, Nishigawa H, Jung HY, Wei W, Suzuki S, Arashida R, Nakata D, Miyata S, Ugaki M, et al. Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat Genet 2004; 36:27-9; PMID:14661021; http://dx.doi.org/10.1038/ng1277
- Tran-Nguyen LTT, Kube M, Schneider B, Reinhardt R, Gibb KS Comparative genome analysis of ‘Candidatus Phytoplasma australiense’ (subgroup tuf-Australia I; rp-A) and “Ca. Phytoplasma asteris” strains OY-M and AY-WB. J Bacteriol 2008; 190:3979-91; PMID:18359806; http://dx.doi.org/10.1128/JB.01301-07
- Hasselbring BM, Jordan LJ, Krause RW, Krause DC. Terminal organelle development in the cell wall-less bacterium Mycoplasma pneumoniae. Proc Natl Acad Sci USA 2006; 103:16478-83; PMID:17062751; http://dx.doi.org/10.1073/pnas.0608051103
- Hafke JB, Ehlers K, Föller J, Höll SR, Becker S, van Bel AJE. Involvement of the sieve-element cytoskeleton in electrical responses to cold shocks. Plant Physiol 2013; 162:707-719.14; PMID:23624858; http://dx.doi.org/10.1104/pp.113.216218
- Rottner K, Stradal TE, Wehland J. Bacteria-host-cell interactions at the plasma membrane: stories on actin cytoskeleton subversion. Dev Cell 2005; 9:3-17; PMID:15992537; http://dx.doi.org/10.1016/j.devcel.2005.06.002
- Radhakrishnan GK, Splitter GA. Modulation of host microtubule dynamics by pathogenic bacteria. Biomol Concepts 2012; 3:571-80; PMID:23585820; http://dx.doi.org/10.1515/bmc-2012-0030
- Galetto L, Bosco D, Balestrini R, Genre A, Fletcher J, Marzachì C. The major antigenic membrane protein of ‘Candidatus Phytoplasma asteris' selectively interacts with ATP synthase and actin of leafhopper vectors. PLoS ONE 2011; 6:e22571; PMID:21799902; http://dx.doi.org/10.1371/journal.pone.0022571
- Boonrod K, Munteanu B, Jarausch B, Jarausch W, Krczal G. An immunodominant membrane protein (Imp) of ‘Candidatus Phytoplasma mali’ binds to plant actin. Mol Plant-Microbe Interact 2012; 25:889-95; PMID:22432876; http://dx.doi.org/10.1094/MPMI-11-11-0303
- Dramsi S, Cossart P. Intracellular pathogens and the actin cytoskeleton. Annu Rev Cell Dev Biol 1998; 14:137-66; PMID:9891781; http://dx.doi.org/10.1146/annurev.cellbio.14.1.137
- Tilney LG, Portnoy DA. Actin filaments and the growth, movements, and spread of the intracellular bacterial parasite Lysteria monocytogens. J Cell Biol 1989; 109:1597-608; PMID:2507553; http://dx.doi.org/10.1083/jcb.109.4.1597
- van Bel AJE. The phloem, a miracle of ingenuity. Plant Cell Environ 2003; 26:125-49; http://dx.doi.org/10.1046/j.1365-3040.2003.00963.x
- Ehlers K Knoblauch M, van Bel AJE. Ultrastructural features of well-preserved and injured sieve elements: minute clamps keep the phloem transport conduits free for mass flow. Protoplasma 2000; 214:80-92; http://dx.doi.org/10.1007/BF02524265
- Swanson MS, Isberg RR Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect Immun 1995; 63:3609-20; PMID:7642298
- Celli J, Tsolis RM. Bacteria, the endoplasmic reticulum and the unfolded protein response: friends or foes? Nature Rev Microbiol 2015; 13:71-82; PMID:25534809; http://dx.doi.org/10.1038/nrmicro3393
- Baruch M, Hertzog BB, Ravins M, Anand A, Cheng CY, Biswas D, Tirosh B, Hanski E. Induction of endoplasmic reticulum stress and unfolded protein response constitutes a pathogenic strategy of groupA streptococcus. Front Cell Infect Microbiol 2014; 4:105; PMID:25136516; http://dx.doi.org/10.3389/fcimb.2014.00105
- van Bel AJE, Ehlers K, Knoblauch M. Sieve elements caught in the act. Trends Plant Sci 2002; 7:126-32; PMID:11906836; http://dx.doi.org/10.1016/S1360-1385(01)02225-7
- Bamunusinghe D, Hemenway CL, Nelson RS, Sanderfoot AA, Ye CM, Silva MA, Payton M, Verchot-Lubicz J. Analysis of potato virus X replicase and TGBp3 subcellular locations. Virology 2009; 393:272-85.27; PMID:19729179; http://dx.doi.org/10.1016/j.virol.2009.08.002
