ABSTRACT
The plant hormone abscisic acid (ABA), a key regulator in many crucial developmental and physiological processes, recruits diverse components into precisely regulated signaling network. We recently discovered that MAPKKK18, an ABA-activated kinase, is regulated by the protein phosphatase type 2C (PP2C) ABI1 and the kinase SnRK2.6, both components of the ABA core signaling pathway. ABI1 acts to inhibit MAPKKK18 kinase activity, but also affects MAPKKK18 protein turnover via the ubiquitin-proteasome pathway. SnRK2.6 kinase also seems to be important for the regulation of MAPKKK18 function. In this review we summarize the mechanisms that are exclusively involved in MAPKKK18 kinase regulation and that ensure specificity in its activation.
Plants use 3 types of effectors to modulate the phosphorylation cascades that are fundamental to the transmission of the ABA signal. The core ABA pathway comprises ABA receptors (the PYR/PYL/RCAR family of START proteins),Citation1-4 together with 2 of these effectors, the group A PP2CsCitation5-9 and the group 2 sucrose nonfermenting-related protein kinases (SnRK2s).Citation10-12 The third type of effector consists of mitogen-activated protein kinases (MAPKs), which, along with other kinases, are also involved in the ABA network,Citation13-16 thus increasing the complexity and diversity of ABA signaling.
Very recently, the MAP3K17/18-MKK3-MPK1/2/7/14 cascade regulated by ABA has been shown to be involved in both senescenceCitation14 and the response to stress.Citation15 Thus, MAP3K17 and MAPKKK18 kinase activity significantly increases during the late stages of ABA treatment.Citation15,16 In addition, a positive correlation is observed between the transcription levels of the genes encoding ABA core complex members and those of the MAP3K17/18 genes. MAP3K17 and MAPKKK18 transcription is abolished in Arabidopsis ABA receptor quadruple pyr1pyl1pyl2pyl4 mutant, as well as in the hab1G246D line (a dominant ABI1-1-like mutation of the HAB1 phosphatase).Citation15 Our latest work focused predominantly on MAPKKK18. For example, we found that MAPKKK18 gene expression is regulated by ABA in specific tissues, e.g. guard cells and root meristem.Citation16 MAPKKK18 expression is also significantly decreased in independent ABI1 knockouts (abi1td and abi1-2), which is consistent with a role for the ABA core pathway in transcriptional activation of MAPKKK18. In fact, MAPKKK18 kinase activity is also increased in the abi1td compared to WT Col-0.Citation16
Apart from transcriptional regulation,Citation15 MAPKKK18 is also strictly controlled at the protein level by ABI1 PP2C. ABI1 not only inhibits MAPKKK18 kinase activity, but is also responsible for targeting MAPKKK18 for degradation by the ubiquitin–proteasome pathway (UPS). Similarly, MAPKKK18 protein accumulation is also affected by ABA, as ABA treatment blocks MAPKKK18 degradation. Because ABA inhibits ABI1 protein phosphatase activity, this suggests that ABA uses ABI1 to regulate turnover of MAPKKK18.Citation16
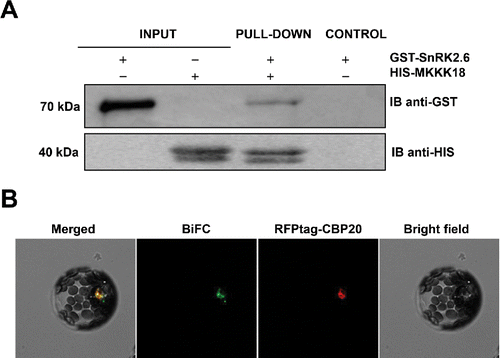
One important element of the ABA core pathway, the SnRK2 kinase, was not deeply investigated in our earlier study, but is most likely also involved in transcriptional regulation of MAPKKK18. Indeed, MAPKKK18 activity is abolished in the snrk2.6 mutant,Citation16 supporting a role for SnRK2.6 in the control of MAPKKK18 expression. We hypothesize that SnRK2.6 phosphorylates a yet unknown transcription factor that drives MAPKKK18 transcription in response to ABA. However, this does not exclude the possibility that SnRK2.6 and MAPKKK18 interact to regulate certain biological processes. To check for a direct interaction between these key kinases, we performed pull-down assays and BiFC analysis and found that, indeed, SnRK2.6 can bind MAPKKK18 (). SnRK2.6 and MAPKKK18 interaction occurred mostly within the nucleus, but minor signals were also observed in the perinuclear cytoplasm. Interestingly, the active form of MAPKKK18 is found within the nucleus, which is where MAPKKK18 also interacts with ABI1. These findings suggest that nuclear localization of MAPKKK18 results in its rapid activation and a consequent resetting of the downstream signaling pathway. The significance of cytoplasm-specific interactions is not yet clear, since only the inactive form of MAPKKK18 should reside in this compartment.
Figure 1. MAPKKK18 interacts with the SnRK2.6 kinase. (A) Pull-down assay to verify the interaction beteween SnRK2.6 and MAPKKK18. Input lines represent total SnRK2.6. Recombinant His- MAPKKK18, pre-coupled to Ni-NTA agarose, was incubated with recombinant GST-SnRK2.6. GST- and His-tagged proteins were detected (IB) using anti-GST and anti-His tag antibodies, respectively. (B) SnRK2.6 and MAPKKK18 proteins interact within the nucleus. Cytoplasmic interaction around the nucleus is present in about 10% of protoplasts. BiFC analysis in Arabidopsis protoplasts transiently expressing full-length SnRK2.6 and MAPKKK18 fused to cECFP or nVenus, respectively. RFPtag-CBP20 was used as both transformation control and marker of nuclear localization. Scale bar: 20 μm.
Based on the proposed role of the ABA core in the regulation of MAPKKK18, we considered possible functions of SnRK2.6 in this regard. Phosphorylation of particular serine/threonine (Ser/Thr) residues by SnRK2.6 might modulate MAPKKK18 protein activity or stability. However, another exciting hypothesis is that MAPKKK18 might regulate SnRK2.6. It will be interesting to investigate whether this interaction is beneficial for both protein partners and whether other relationships might exist.
In conclusion, current results demonstrate that regulation of signal transduction via the MAPKKK18 cascade is complex. Specificity of MAPKKK18 regulation is achieved by 2 exclusive mechanisms: one mechanism ensures activation of MAPKKK18 by ABA through coordinated MAPKKK18 gene expression regulated by the core ABA pathway; the second, more complex mechanism for MAPKKK18 signal inactivation is based on proteasomal degradation, which is regulated by ABA via the ABA core pathway. These mechanisms demonstrate the dependence of MAPKKK18 regulation on the components of the ABA core pathway and show the importance of MAPKKK18 in signal transduction. Identification of all the players in the MAPKKK18 network that affect its activation or inactivation, as well as the identification of MAPKKK18 cascade substrates, is a major challenge in the near future.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from the National Science Center (No. DEC-2012/05/B/NZ3/00352 and 2015/17/N/NZ3/00532).
References
- Meyer K, Leube MP, Grill E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 1994; 264(5164):1452-5; PMID:8197457; http://dx.doi.org/10.1126/science.8197457
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009; 324:1064-8; PMID:19407143; http://dx.doi.org/10.1126/science.1172408
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodriguez A, Chow Tsz-fung, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009; 324(5930):1068-71; PMID:19407142; http://dx.doi.org/10.1126/science.1173041
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell D F, Cutler SR, Chory J, et al. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J 2010; 61:290-9; PMID:19874541; http://dx.doi.org/10.1111/j.1365-313X.2009.04054.x
- Gosti F, Beaudoin N, Serizet C, Webb A, Vartanian N, Giraudat J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 1999; 11:1897-910; PMID:10521520; http://dx.doi.org/10.1105/tpc.11.10.1897
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 2006; 140:115-26; PMID:16339800; http://dx.doi.org/10.1104/pp.105.070128
- Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T. ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 2007; 50:935-49; PMID:17461784; http://dx.doi.org/10.1111/j.1365-313X.2007.03107.x
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Márquez J A, Cutler SR, Rodriguez PL. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 2009; 60:575-88; PMID:19624469; http://dx.doi.org/10.1111/j.1365-313X.2009.03981.x
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim T, Santiago J, Flexas J, Schroeder JI, Rodriguez PL. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol 2009; 150:1345-55; PMID:19458118; http://dx.doi.org/10.1104/pp.109.137174
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Yasushi H, Shinozaki K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci U S A 2009; 106(41):17588-93; PMID:19805022; http://dx.doi.org/10.1073/pnas.0907095106
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 2009; 21:3170-84; PMID:19855047; http://dx.doi.org/10.1105/tpc.109.069179
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 2010; 61:65179; PMID:20192755; http://dx.doi.org/10.1146/annurev-arplant-042809-112122
- Liu Y. Roles of mitogen-activated protein kinase cascades in ABA signaling. Plant Cell Rep 2012; 31(1):1-12; PMID:21870109; http://dx.doi.org/10.1007/s00299-011-1130-y
- Matsuoka D, Yasufuku T, Furuya T, Nanmori T. An abscisic acid inducible Arabidopsis MAPKKK, MAPKKK18 regulates leaf senescence via its kinase activity. Plant Mol Biol 2015; 87:565-75; PMID:25680457; http://dx.doi.org/10.1007/s11103-015-0295-0
- Danquah A, de Zélicourt A, Boudsocq M, Neubauer J, Frei dit Frey N, Leonhardt N, Pateyron S, Gwinner F, Tamby JP, Ortiz-Masia D, et al. Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J 2015; 82:232-44; PMID:25720833; http://dx.doi.org/10.1111/tpj.12808
- Mituła F, Tajdel M, Cieśla A, Kasprowicz-Maluśki A, Kulik A, Babula-Skowrońska D, Michalak M, Dobrowolska D, Sadowski J, Ludwików A. Arabidopsis ABA-activated kinase MAPKKK18 is regulated by protein phosphatase 2C ABI1 and the ubiquitin proteasome pathway. Plant Cell Physiol 2015; 56(12):2351-67; Oct 6 pii: pcv146; PMID:26443375; http://dx.doi.org/10.1093/pcp/pcv146
