ABSTRACT
As obligate photosynthetic organisms plants are particularly exposed to the damaging effects of excess light and ultraviolet wavelengths, which can impact genome and epigenome dynamics by inducing DNA sequence and chromatin alterations. DNA DAMAGE-BINDING PROTEIN 2 (DDB2) is the main factor involved in the recognition of UV-induced DNA lesions during Global Genome Repair (GGR) in mammals and in plants.Citation1 In a recent study we reported that, in Arabidopsis, loss of DDB2 function alters DNA methylation patterns at many repeat loci and protein coding genes. We demonstrated that DDB2 acts in a complex with ARGONAUTE 4 (AGO4) to control de novo DNA methylation via the modulation of the local abundance of 24-nt small interfering RNAs (siRNAs). In addition, we found that DDB2 negatively regulates the expression of REPRESSOR OF SILENCING 1 (ROS1), a primary factor required for active DNA demethylation. Here we report that depletions of cognate GGR factors also lead to alterations of DNA methylation profiles at particular loci. Taken together, these findings reveal an interplay between GGR factors and DNA methylation patterns.
Abbreviations
| AGO4 | = | ARGONAUTE 4 |
| CEN2 | = | CENTRIN2 |
| DDB2 | = | DNA DAMAGE-BINDING PROTEIN 2 |
| DMRs | = | Differentially Methylated Regions |
| MSH1 | = | MutS HOMOLOG 1 |
| RdDM | = | RNA-directed DNA methylation |
| ROS1 | = | REPRESSOR OF SILENCING 1 |
| siRNAs | = | short interfering RNAs |
| TE | = | Transposable Element |
Dynamic changes in the epigenome have increasingly been recognized to be important in plant development and for the response of plants to environmental stress.Citation2 DNA methylation (5-methylcytosine) is one epigenetic mark that is associated with compacted chromatin structure, the regulation of gene expression, as well as being important for transposon (TE) silencing and genome imprinting.Citation3 In plants, methylation of cytosine in DNA can occur in both symmetric (CG and CHG) and asymmetric (CHH) contexts, where H is either A, T or C, allowing a multitude of distinct 5-methylcytosine patterns.Citation3 DNA methylation is a dynamic modification, which is (at times) established de novo and which must be maintained, but which can be lost via active or passive DNA demethylation.Citation3 Together, these dynamics change chromatin structure, influencing the flexibility and stability of genomes.
Active DNA demethylation is a DNA repair-based process affecting cytosine methylation in all sequence contexts.Citation4 In Arabidopsis, loss of function of the Mismatch Repair factor MutS HOMOLOG 1 (MSH1) leads to heritable alterations of the DNA methylation landscape.Citation5 Thus it seems reasonable that specific factors at the nexus of DNA repair and DNA methylation could impact genome integrity and the epigenome, thereby enhancing the plasticity of these biological information systems. Direct interconnections between DNA repair processes and DNA methylation/demethylation machineries should thus be thoroughly investigated.
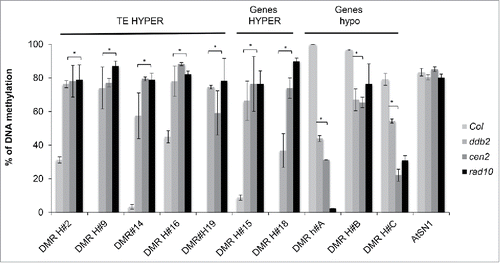
Given that GGR corrects UV-induced DNA lesions and that DDB2 shapes the DNA methylation landscape,Citation6 it was tempting to evaluate the putative role of known GGR factors on DNA methylation. For this we extended our analysis to a mutant affecting CENTRIN2 (CEN2), which is part of the XPC complex (Xeroderma pigmentosum, complementation group C) that recognizes bulky DNA adducts,Citation7,8 and to a mutant affecting RAD10, which is an endonuclease involved in the dual excision of photoproducts.Citation1 Using the methylation-dependent restriction enzyme, McrBC, coupled to qPCR, we found that both cen2 and rad10 mutant plants exhibit DNA methylation alterations similar to those observed in several previously identified ddb2-induced Differentially Methylated RegionsCitation6 (DMRs; ). These data strongly suggest that GGR factors directly or indirectly shape the DNA methylation landscape, at particular loci, even in the absence of genotoxic stress.
Figure 1. DNA methylation in GGR-deficient plants. DNA methylation levels of WT (Col), cen2 and rad10 plants determined by McrBC-qPCR at several representative ddb2-induced Differentially Methylated Regions (DMRs). Data are presented as percentage of methylation (±SD ) and are representative of 3 biological replicates. The methylated transposable retro-element, SINE like element 1 (AtSN1), was used as control. t-test * p < 0.01 compared to WT (Col).
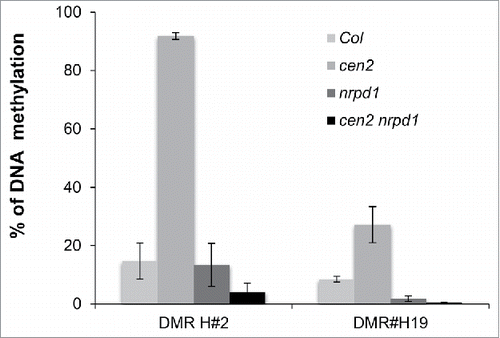
To assess whether the gain of DNA methylation over TEs regions observed in cen2 plants depends on the RNA-directed DNA methylation (RdDM) pathway, as was observed in ddb2 mutant plants, we tested cen2-identified hyper-DMRs, overlapping with 24-nt siRNAs, in plants simultaneously mutant for CEN2 and the largest subunit of RNA polymerase IV (NRPD1), which is a key factor in RdDM. McrBC-qPCR analyses showed that cen2-induced hypermethylation was suppressed in cen2 nrpd1 double mutant plants (), indicating that RdDM is responsible for the DNA hypermethylation observed at particular TEs, when GGR is compromised.
Figure 2. RdDM-dependent gain of DNA methylation in GGR-deficient plants. Genetic interaction between cen2 and nrpd1. Percentage of DNA methylation in Col, cen2, nrpd1 and in cen2-nrpd1 plants for 2 representative TEs overlapping 24-nt siRNAs. Data are presented as percentage of methylation (±SD) determined by McrBC-qPCR and are representative of 3 biological replicates.
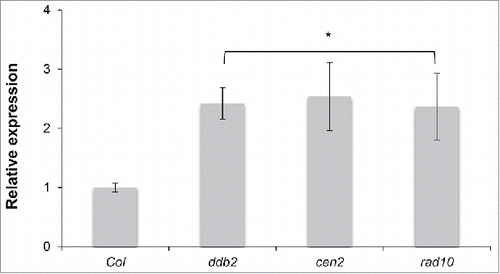
A portion of the DNA methylation changes observed in ddb2 mutant plants resulted from changes caused by expression in genes involved in active DNA demethylation.Citation6 Transcript levels of ROS1 were tested by RT-qPCR in cen2 and rad10 mutant plants. Interestingly, elevated ROS1 mRNA levels were also detected in cen2 and rad10 plants (), showing that GGR factors control expression of this particular DNA demethylase and likely the DNA methylation pattern of their genomic targets.
Figure 3. ROS1 expression levels in GGR-deficient plants. RT-qPCR analysis of transcript levels (±SD ) of genes encoding the DNA glycosylase ROS1 in WT (Col), ddb2, cen2 and rad10 plants. t-test * p < 0.01 compared to WT (Col).
DDB2 is the first detection factor acting during the GGR process acting upstream of CEN2, and functions to stabilize XPC on chromatin.Citation7 The presence of the XPC-CEN2 complex was shown to competitively inhibit DDB2 polyubiquitination and its subsequent degradation by enhancing its retention on chromatin.Citation9 Therefore, the enrichment of DDB2 on chromatin in cen2 mutant plants may be responsible for DNA methylation changes detected in these plants. More generally, it is likely that GGR factors or other specific effectors regulating DDB2 homeostasis/dynamics (e.g. CULLIN4, ATR: Ataxia telangiectasia and Rad3 relatedCitation10) also play a regulatory role in the control of DNA methylation.
This study reports an essential role for several GGR factors in shaping the Arabidopsis DNA methylation landscape, at particular loci, independently of exogenous induction of DNA damage. Therefore, it is tempting to speculate that the GGR machinery controls, genome wide, the balance between active DNA demethylation and de novo DNA methylation to cooperatively maintain genome and epigenome integrity. Such effects could occur indirectly via post-translational regulation of key factors (i.e., DDB2) or directly via recognition of yet uncharacterized DNA/chromatin features. In the future, it would be essential to confirm, genome wide, that CEN2, RAD10 but also that other GGR factors play a role in shaping DNA methylation landscape and to decipher the molecular mechanisms regulating the interplay between the GGR and DNA methylation pathways.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Schärer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol 2013; 5:a012609; PMID:24086042; http://dx.doi.org/10.1101/cshperspect.a012609
- Chinnusamy V, Zhu JK. Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 2009; 2:133-39; PMID:19179104; http://dx.doi.org/10.1016/j.pbi.2008.12.006
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 2010; 11:204-20; PMID:20142834; http://dx.doi.org/10.1038/nrg2719
- Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet 2009; 43:143-66; PMID:19659441; http://dx.doi.org/10.1146/annurev-genet-102108-134205
- Virdi KS, Laurie JD, Xu YZ, Yu J, Shao MR, Sanchez R, Kundariya H, Wang D, Riethoven JJ, Wamboldt Y, Arrieta-Montiel MP, Shedge V, Mackenzie SA. Arabidopsis MSH1 mutation alters the epigenome and produces heritable changes in plant growth. Nat Commun 2015; 6:6386; PMID:25722057; http://dx.doi.org/10.1038/ncomms7386
- Schalk C, Drevensek S, Kramdi A, Kassam M, Ahmed I, Cognat V, Graindorge S, Bergdoll M, Baumberger N, Heintz D, Bowler C, Genschik P, Barneche F, Colot V, Molinier J. DNA DAMAGE BINDING PROTEIN 2 (DDB2) shapes the DNA methylation landscape. Plant Cell 2016; In press, pii: tpc.00474; PMID:27531226; http://dx.doi.org/10.1105/tpc.16.00474
- Araki M, Masutani C, Takemura M, Uchida A, Sugasawa K, Kondoh J, Ohkuma Y, Hanaoka F. Centrosome protein centrin 2/caltractin 1 is part of the Xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J Biol Chem 2001; 276:18665-72; PMID:11279143; http://dx.doi.org/10.1074/jbc.M100855200
- Liang L, Flury S, Kalck V, Hohn B, Molinier J. CENTRIN2 interacts with the Arabidopsis homolog of the human XPC protein (AtRAD4) and contributes to efficient synthesis-dependent repair of bulky DNA lesions. Plant Mol Biol 2006; 1-2:345-56; PMID:16786311; http://dx.doi.org/10.1007/s11103-006-0016-9
- Matsumoto S, Fischer ES, Yasuda T, Dohmae N, Iwai S, Mori T, Nishi R, Yoshino K, Sakai W, Hanaoka F, Thomä NH, Sugasawa K. Functional regulation of the DNA damage-recognition factor DDB2 by ubiquitination and interaction with Xeroderma pigmentosum group C protein. Nucleic Acids Res 2015; 43:1700-13; PMID:25628365; http://dx.doi.org/10.1093/nar/gkv038
- Molinier J, Lechner E, Dumbliauskas E, Genschik P. Regulation and role of Arabidopsis CUL4-DDB1A-DDB2 in maintaining genome integrity upon UV stress. PLoS Genet 2008; 4:e1000093; PMID:18551167; http://dx.doi.org/10.1371/journal.pgen.1000093
