ABSTRACT
Sorghum is a short day plant with strong photoperiod response and its cultivation for grain in temperate regions necessitated the development of photoperiod insensitive mutants that can flower rapidly in the long days of summer. Wild type genotypes grow vegetatively in summer accumulating significant biomass before floral transition ensues during the shorter days of fall. Thus, photoperiod insensitive mutants are grown for grain production while photoperiod sensitive wild type genotypes are grown for forage and biomass feedstock production in the United States. However, the molecular mechanism of photoperiod response and floral transition is poorly understood in sorghum. We have previously reported 3 FLOWERING LOCUS T homologues (SbFT1, SbFT8 and SbFT10) that serve as the ultimate mediators of photoperiod response and floral transition, but more work remains to be done to clearly define the molecular function of the upstream regulatory factors. One of the major QTL that accounts for 85% of the flowering time variation, which was reported to be encoding the PRR37 protein is now debated to be encoding the SbFT12 protein, raising further questions as to how SbFT12 may regulate sorghum florigens. Further molecular analyses will uncover the true nature of the day length sensors in sorghum and the mechanisms of their interactions with florigens to modulate photoperiod dependent vegetative growth and floral transition.
Sorghum (Sorghum bicolor L. Moench) is a multipurpose crop grown for food, feed and fuel in tropical, subtropical, temperate, and semi-arid regions of the world. It is the fifth most important cereal worldwide after maize, wheat, rice, and barley. As a short day tropical species, sorghum exhibits substantial photoperiod sensitivity and this has important implication in its agronomy in temperate regions. Sorghum flowers rapidly in short day photoperiodsCitation1 but the extended light period in temperate regions allows more photosynthesis and extensive vegetative growth during the warm months of summer. In the United States and other temperate regions, breeders have selected for mutants that flower rapidly independent of day length for grain production while keeping the photoperiod sensitivity trait in forage and biomass producing genotypes to allow more biomass accumulation in the long summer days. Thus, whether a given sorghum germplasm is grown for grain or biomass production in temperate regions depends on its photoperiod response behavior. However, the molecular mechanism of photoperiod response in sorghum and its relationship to developmental programs is poorly understood. Flowering time is obviously a key feature of photoperiod response. While photoperiod insensitive genotypes flower early irrespective of day length, photoperiod sensitive genotypes flower rapidly in short days (SDs) but flower very late in long days (LDs). But floral transition is not the only trait affected by photoperiod. Other developmental parameters including plant height, stem thickness and leaf size differ markedly under LD and SD growth conditions. Vegetative growth is significantly reduced but floral transition is accelerated under SD conditions. However, the reduction in vegetative growth does not appear to be due only to rechanneling of resources to reproductive growth because photoperiod insensitive genotypes grown under SDs also accumulate less biomass compared to those grown in LDs although flowering time is not significantly different between LDs and SDs. For example, the photoperiod insensitive grain sorghum genotype BTx623 flowers in a comparable time under LD and SD conditions but vegetative growth reduction in SDs is similar to that of photoperiod sensitive genotypes sweet sorghum Theis and forage sorghum FS000991 (), both of which flower early in SDs but very late in LDs. This suggests that the presence of florigen or the flowering signal per se may not be the only cause for the reduced growth in SDs. At least, if florigen is the cause, it should be interacting with other factors to prevent growth specifically in SDs.
Figure 1. Growth responses of sorghum genotypes to photoperiod. Vegetative growth characteristics of photoperiod in sensitive (A) and sensitive (B and C) genotypes of sorghum grown for 6 weeks under long day (LD) with 16 hours of light period or short day (SD) with 8 hours of light period.

To quantify the effect of inductive SD treatment on overall plant growth and development, we systematically recorded leaf number, size, width, length, stem thickness (diameter), internode length and plant height of sweet sorghum Theis variety as this variety is extremely responsive to photoperiod variation. We found that the leaf number of Theis never exceeds 9 at 8 hour SD photoperiods whereas, under LD conditions the average leaf number reaches to 19. In general, the leaf length and width of Theis were reduced by 2 to 4-fold, and plant height, internode length and stem thickness were reduced by 2 to 5-fold under SD conditions (). Overall developmental condition of Theis under SD is characterized by short period growth time with early flowering and maturity, but dramatically reduced total biomass accumulation. One major reason for this could be that in SDs, there is little time for solar energy harvest and photosynthesis is limiting. However, this is in sharp contrast to the LD plant Arabidopsis thaliana, where SD not only delays flowering but also increases leaf number and leaf size. Citation2 Photosynthesis is the source of building blocks for biomass accumulation in both SD and LD plants but it is unclear why it should be limiting in the SD growth of SD plants but not in that of LD plants. It can be argued that since the LD plants take more time to flower in SDs, photosynthesis is compensated over an extended period of growth. But this does not explain why leaf size per se is bigger in LD plants but smaller in SD plants under SD conditions. It is likely that some components of the floral transition pathway and the photoperiod response regulators interact in some way to positively or negatively affect plant growth and development specific to the photoperiod requirements of plants. Consistent with this, several Arabidopsis long day flowering pathway mutants flower with even more and larger leaves in LDs than their respective wild types in SDs.Citation2,3 The challenge for sorghum is to identify these factors that interact with photoreceptors and florigens to specifically alter plant growth depending on day length.
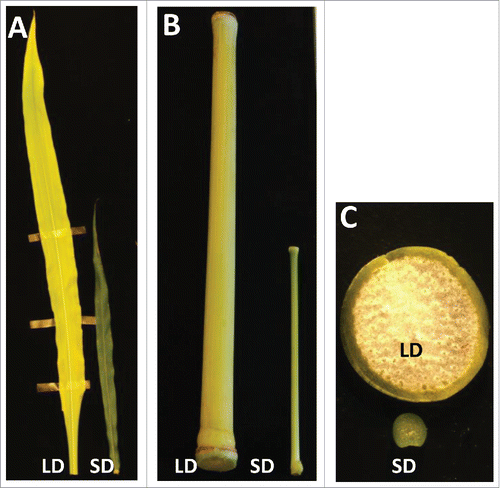
Figure 2. Morphology of sweet sorghum Theis genotype,grown under long day (LD) and short day (SD) conditions. Leaves (A) and internodes (B) from equivalent positions at maturity are shown from plants grown under long days (LD) or short days (SD). (C), Cross section of stems from equivalent position grown under LD or SD conditions.
In rice, another SD plant, the promoter and repressor activities of Early heading date 1 (Ehd1) and Grain number, plant height, and heading date 7 (Ghd7), respectively, enable to control the rice florigen Hd3a transcription.Citation4 Both Ehd1Citation5 and Ghd7Citation6 appear to be monocot specific and their effects seem to widely vary between SD and LD plants. While rice Ghd7 is a repressor of flowering in LDs, its homolog VRN2 confers winter habit in the LD plant wheat.Citation7 Other common factors such as Heading date 1 (Hd1) may have opposite effects under SD and LD conditions. Citation8 Arabidopsis CONSTANS (CO) is a floral activator, Citation9 while its rice ortholog Hd1 promotes flowering in inductive SDs but represses flowering in non-inductive LDs through interacting with Ghd7 to repress Ehd1.Citation10 Thus, if factors like Hd1 for example, have an inherent effect in plant size, their activation or repression activity could contribute to control of vegetative growth in addition to floral transition in response to day length. The situation is even grimmer in sorghum as much is not known about the mechanism of photoperiod response or floral transition. Photoreceptors and homologues of Ehd1 and Ghd7 have been reported in sorghumCitation11,12 but their mechanistic actions on vegetative growth and/or floral transition have not been comprehensively understood.
Eleven non-overlapping QTLs for floral transition have been described in sorghumCitation13,14 of which 6 (ma1 – ma6) have been used as maturity loci in breeding programs.Citation1,15-17 Photoperiod sensitivity and delayed flowering in LDs is conferred by additive effects of dominant genes at each loci. For example, a genotype Ma1, Ma2, Ma3 is more photoperiod sensitive and delayed flowering in LDs than ma1, ma2, Ma3 genotype. Ma1 and Ma3 have the most significant effects and have been considered to account together for most of the differences in photoperiod sensitivity. Ma1 was reported to encode a pseudo response regulator floral repressor PRR37,Citation18 homologous to barley Ppd-H1, which is a promoter of flowering in LDs.Citation19 A recent report, however, associated Ma1 to SbFT12 rather than SbPRR37.Citation20 Ma3 was reported to encode Phytochrome B (PHYB)Citation21 although definitive proof by mutant complementation is still lacking. Whether Ma1 encodes SbPRR37 or SbFT12, the long held view is that Ma1 is activated by PHYB in LDs and represses transcription of florigens to delay flowering.Citation18 We have recently reported that sorghum has at least 3 potential florigen encoding genes, SbFT1, SbFT8 and SbFT10 that activate floral transition.Citation22 These 3 genes are expressed in the leaves of photoperiod insensitive genotypes at the floral transition stage irrespective of day length but induced by SDs in photoperiod sensitive genotypes, suggesting that sorghum florigens are the ultimate mediators of photoperiod response. However, whether sorghum florigens directly respond to the light signal and be able to measure day length or simply respond to upstream repressors is unclear. The fact that in the classical sorghum mutants, 38M (ma1, ma2, ma3R) flowers earlier in LDs and has more florigen encoding transcripts than 44M (ma2, ma3R) and this intern flowers earlier and has more transcripts than 100M (Ma1, Ma2, Ma3, Ma6) Citation22 favors the hypothesis that florigens are repressed by photoperiod sensors rather than measure day length themselves. Nevertheless, this needs to be demonstrated directly. Importantly, if Ma1 is indeed SbFT12, it will be very interesting to investigate how SbFT12 represses the downstream florigens and whether it is capable of directly sensing day length.
On the other hand, with 3 putative florigen encoding genes, sorghum floral activation appears to combine the homologues of rice Hd3a (SbFT1) and maize ZCN8 (SbFT8 and SbFT10). The relative contributions of these genes to sorghum florigen is not known although all the 3 genes have properties of a florigen, including activation of flowering in transgenic Arabidopsis. Why sorghum needed 3 florigens more than rice and maize is unclear. What also remains unclear is the role of physical protein-protein interaction in determining florigenic potential. Arabidopsis FT directly interacts with FD to activate AP1Citation23 while rice Hd3a interacts with OsFD1 indirectly through a 14-3-3 protein.Citation24 In sorghum, SbFT1 directly interacts with SbFD1, Sb14-3-3, SbFT8, and SbFT10 in yeast-to-hybrid (Y2H) and Bimolecular fluorescence complementation (BiFC) assaysCitation22 while SbFT8 and SbFT10 directly interact with Sb14-3-3 but not with SbFD1 similar to rice Hd3a. But there is no evidence at this stage to suggest that direct interaction may be more important than indirect interaction for functional significance in floral activation as even the antagonistic floral repressor Arabidopsis TFL1 interacts with FD to repress flowering.Citation25 Further molecular analyses would be needed to understand the mechanism with which reversion of FT induction by LDs, memory of exposure to SDs, as well as de-repression of axillary meristem during photoperiod switching are achieved in sorghum, which will provide novel mechanistic insight in the response of sorghum developmental programs to environmental signals. Extensive experiments with mutants or reduced expression lines will be required to quantitatively determine the contribution of each of the 3 SbFT genes to sorghum flowering and photoperiod response. It will also be important to investigate how these 3 SbFTs and other floral promoters are regulated by the circadian clock (GI and CO), photoreceptors (PHYB, PHYC, Cryptochromes) and transcriptional regulators for a comprehensive understanding of the molecular mechanisms of signaling connections between photoperiod response, plant development, biomass accumulation and floral transition in sorghum.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
This work was supported by the National Institute of Food and Agriculture, US. Department of Agriculture, under award number 2013-69005-21284.
References
- Quinby J, Karper R. The inheritance of three genes that influence time of floral initiation and maturity date in milo. Agron J 1945; 37:916-36; http://dx.doi.org/10.2134/agronj1945.00021962003700110006x
- Amasino R. Seasonal and developmental timing of flowering. Plant J 2010; 61:1001-13; PMID:20409274; http://dx.doi.org/10.1111/j.1365-313X.2010.04148.x
- Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T. Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol 2015; 66:441-64; PMID:25534513; http://dx.doi.org/10.1146/annurev-arplant-043014-115555
- Itoh H, Nonoue Y, Yano M, Izawa T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat Genet 2010; 42:635-8; PMID:20543848; http://dx.doi.org/10.1038/ng.606
- Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 2004; 18:926-36; PMID:15078816; http://dx.doi.org/10.1101/gad.1189604
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 2008; 40:761-7; PMID:18454147; http://dx.doi.org/10.1038/ng.143
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 2004; 303:1640-4; PMID:15016992; http://dx.doi.org/10.1126/science.1094305
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the arabidopsis flowering time gene CONSTANS. Plant Cell 2000; 12:2473-83; PMID:11148291; http://dx.doi.org/10.1105/tpc.12.12.2473
- Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995; 80:847-57; PMID:7697715; http://dx.doi.org/10.1016/0092-8674(95)90288-0
- Nemoto Y, Nonoue Y, Yano M, Izawa T. Hd1,a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J 2016; 86:221-33; PMID:26991872; http://dx.doi.org/10.1111/tpj.13168
- Yang S, Murphy R, Morishige D, Klein P, Rooney W. Sorghum phytochrome B inhibits flowering in long days by activating expression of SbPRR37 and SbGHD7, repressors of SbEHD1, SbCN8 and SbCN12. PLoS ONE 2014; 9:e105352; PMID:25122453; http://dx.doi.org/10.1371/journal.pone.0105352
- Murphy R., et al. Ghd7 (Ma6) represses flowering in long days: a key trait in energy sorghum hybrids. PLoS One 2014; 9:e105352; PMID:25122453; http://dx.doi.org/10.1371/journal.pone.0105352
- Zhang D, Kong W, Robertson J, Goff VH, Epps E, Kerr A, Mills G, Cromwell J, Lugin Y, Phillips C,, et al. Genetic analysis of inflorescence and plant height components in sorghum (Panicoidae) and comparative genetics with rice (Oryzoidae). BMC Plant Biol 2015; 15:107; PMID:25896918; http://dx.doi.org/10.1186/s12870-015-0477-6
- Zhang D, Guo H, Kim C, Lee TH, Li J, Robertson J, Wang X, Wang Z, Paterson AH. CSGRqtl, a comparative quantitative trait locus database for saccharinae grasses. Plant Physiol 2013; 161:594-9; PMID:23370713; http://dx.doi.org/10.1104/pp.112.206870
- Quinby J. The genetic control of flowering and growth in sorghum. Adv Agron 1973; 25:125-62; http://dx.doi.org/10.1016/S0065-2113(08)60780-4
- Vanderlip RL, Reeves HE. Growth stages of sorghum. Agronomy J 1972; 64:13-17; http://dx.doi.org/10.2134/agronj1972.00021962006400010005x
- Quinby J. Fourth maturity gene locus in sorghum. Crop Sci 1966; 6:516-8; http://dx.doi.org/10.2135/cropsci1966.0011183X000600060005x
- Murphy R, Klein RR, Morishige DT, Brady JA, Rooney WL, Miller FR, Dugas DV, Klein PE, Mullet JE. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc Natl Acad Sci USA 2011; 108:16469-74; PMID:21930910; http://dx.doi.org/10.1073/pnas.1106212108
- Turner A, Beales J, Faure S, Dunford R, Laurie D. The pseudoresponse regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 2005; 310:1031-4; PMID:16284181; http://dx.doi.org/10.1126/science.1117619
- Cuevas HE, Zhou C, Tang H, Khadke PP, Das S, Lin YR, Ge Z, Clemente T, Upadhyaya HD, Hash CT, et al. The evolution of photoperiod-insensitive flowering in sorghum, a genomic model for panicoid grasses. Mol Biol Evol 2016; 33:2417-28; PMID:27335143; http://dx.doi.org/10.1093/molbev/msw120
- Childs K, Miller FR, Cordonnier-Pratt MM, Pratt LH, Morgan PW, Mullet JE. The sorghum photoperiod sensitivity gene, Ma3, encodes a phytochrome B. Plant Physiol 1997; 113:611-9; PMID:9046599; http://dx.doi.org/10.1104/pp.113.2.611
- Wolabu TW, Zhang F, Niu L, Kalve S, Bhatnagar-Mathur P, Muszynski MG, Tadege M. Three FLOWERING LOCUS T-like genes function as potential florigens and mediate photoperiod response in sorghum. New Phytol 2016; 210:946-59; PMID:26765652; http://dx.doi.org/10.1111/nph.13834
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005; 309:1052-6; PMID:16099979; http://dx.doi.org/10.1126/science.1115983
- Taoka K, et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 2011; 476:332-7; PMID:21804566; http://dx.doi.org/10.1038/nature10272
- Hanano S, Goto K. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 2011; 23:3172-84; PMID:21890645; http://dx.doi.org/10.1105/tpc.111.088641
