ABSTRACT
Essential genes are usually less likely to be lost during evolution, whereas dispensable genes are lost more frequently. Integrating sacred lotus and other plant microRNA (miRNA) data, we found different ancient miRNA families that arose before eudicot radiation exhibit different evolutionary trajectories. Those ancient miRNA families with higher copy and target numbers, and older age are more likely to be retained in plant descendants and more conserved in (hairpin-structured) miRNA gene sequences. Interestingly, a large portion of the well conserved miRNA families in plant lineages can target transcription factors (TFs). Also, we found miRNA families that target TFs are preferentially retained after sacred lotus genome duplication. In this article, we provide some points to discuss why miRNA families that regulate TFs are more likely to be preserved in plants.
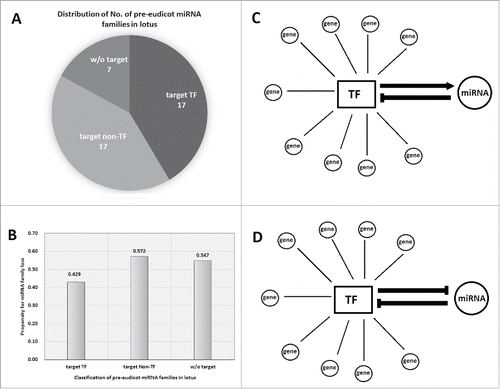
Transcription factors (TFs) are among the largest gene families in plants.Citation1 A great number of molecular genetic and omic studies identified diverse microRNAs (miRNAs) that can regulate TFs through translational repression or cleavage of mRNAs.Citation2 However, other large gene families such as receptor-like kinases (RLKs) or ubiquitins are with few miRNAs being reported to regulate them.Citation3 In the lotus genome, 1,476/26,685 (5.5%) protein-coding genes encode TFs, while 83/249 (33.3%) miRNAs with target gene(s) are able to target TFs. More intriguingly, 17/34 (50.0%) ancient miRNA families, arose before eudicot radiation, with target(s) in lotus can regulate TFs (). These suggest that TF-related miRNAs were biasedly preserved during the plant evolution.Citation4 But why such bias in TF-related miRNAs? First, this group of target genes of miRNAs, TFs, are at the hubs of gene regulatory networks; second, TFs are dosage-sensitive; third, loss-of-function mutations of TFs usually produce severe phenotypes of plants.Citation5-7 For example, miR396, which appeared in the MRCA (most recent common ancestor) of vascular plants, has important function in regulation of leaf growth via targeting the TF family GRFs.Citation4,8 Moreover, studies found that different TFs and miRNAs are often interlaced in the transcriptional networks (). For example, upon phosphorus deficiency in Arabidopsis, expression of the TF, PHR1, can induce transcription of miR399, which can subsequently target PHO2, to regulate phosphorous homeostasis.Citation5 Intriguingly, transcription of some MIRNAs (miRNA genes) requires specific TFs to bind MIRNA promoters to activate pri-miRNA transcription, and these TFs also happen to be their targets, and therefore they form a negative feedback loop (). For example, in Arabidopsis, miR160/miR167 and ARFs form this feedback loop to regulate different developmental processes including adventitious root formation.Citation9 Similarly, miR156-SPLs, miR172-AP2 also form as 2 negative feedback loops in regulation of developmental timing in Arabidopsis.Citation10 Moreover, the miRNA-regulated TFs usually have multiple duplicates in the plant genome and preserve the same miRNA-binding sites. Therefore a single miRNAs usually regulate multiple closely related TFs, making it at the hub of miRNA-TF-genes networks (). All these examples indicate that mutually regulatory relation between TFs and miRNAs potentially enhance the functional essentiality of TF-related miRNAs.
Figure 1. Pre-eudicot miRNA families that regulate transcription factors (TFs) are well-conserved in plants. (A) Distribution of pre-eudicot miRNA families that target different targets. (B) Pre-eudicot miRNA families that regulate TFs are less likely to be lost in plant. (C,D) Two models of miRNA-TF regulatory relationships.
In molecular function, both miRNAs and TFs function as trans-acting factors to regulate target gene at mRNA level. Thus, miRNAs may assist TFs to improve the efficiency and accuracy of gene regulation at RNA level. Other large gene families, for instance, RLKs or ubiquitins, mainly regulate cell response or growth via signaling transduction at protein modification level, which proceed much faster than regulation by transcription and translation.Citation11-13 Assuming that, during the brassinosteroid signaling, a miRNA was evolved to regulate the mRNA of a RLK (such as BRI1) and the RLK can regulate miRNA level through signaling transduction via downstream TFs (for example BES1), this assumed regulatory loop would be less efficient than a peptide or protein to directly modify the signaling of the RLK or a miRNA to regulate the mRNA level of the downstream TF.Citation14,15 Taken together, these points might be the reasons why we observed more TF-associated miRNAs than other functional miRNAs being conserved in the plant genomes.
Disclosure of potential conflicts of interest
The authors confirmed that there is no potential conflicts of interest.
Funding
This work was supported by The Knowledge Innovation Project of the Chinese Academy of Sciences (grant number Y455421Z02) and National Natural Science Foundation of China (grant number 31471899).
References
- Perez-Rodriguez P, Riano-Pachon DM, Correa LG, Rensing SA, Kersten B, Mueller-Roeber B. PlnTFDB: updated content and new features of the plant transcription factor database. Nucl Acid Res 2010; 38:D822-7; PMID:19858103; http://dx.doi.org/10.1093/nar/gkp805
- Meng Y, Shao C, Wang H, Chen M. The regulatory activities of plant microRNAs: a more dynamic perspective. Plant Physiol 2011; 157:1583-95; PMID:22003084; http://dx.doi.org/10.1104/pp.111.187088
- Shi T, Huang H, Sanderson MJ, Tax FE. Evolutionary dynamics of leucine‐rich repeat receptor‐like kinases and related genes in plants: A phylogenomic approach. J Integr Plant Biol 2014; 56:648-62; http://dx.doi.org/10.1111/jipb.12188
- Shi T, Wang K, Yang P. The evolution of plant microRNAs—insights from a basal eudicot sacred lotus. Plant J 2016; PMID:27743419; http://dx.doi.org/10.1111/tpj.13394
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 2007; 449:1053-7; PMID:17960244; http://dx.doi.org/10.1038/nature06206
- Barah P, B NM, Jayavelu ND, Sowdhamini R, Shameer K, Bones AM. Transcriptional regulatory networks in Arabidopsis thaliana during single and combined stresses. Nucl Acid Res 2016; 44:3147-64; PMID:26681689; http://dx.doi.org/10.1093/nar/gkv1463
- Guo Z, Fujioka S, Blancaflor EB, Miao S, Gou X, Li J. TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 2010; 22:1161-73; PMID:20435901; http://dx.doi.org/10.1105/tpc.109.069203
- Debernardi JM, Rodriguez RE, Mecchia MA, Palatnik JF. Functional specialization of the plant miR396 regulatory network through distinct microRNA - target interactions. PLoS Genet 2012; 8:e1002419; PMID:22242012; http://dx.doi.org/10.1371/journal.pgen.1002419
- Gutierrez L, Bussell JD, Păcurar DI, Schwambach J, Păcurar M, Bellini C. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 2009; 21:3119-32; PMID:19820192; http://dx.doi.org/10.1105/tpc.108.064758
- Wu G, Park MY, Conway SR, Wang J-W, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009; 138:750-9; PMID:19703400; http://dx.doi.org/10.1016/j.cell.2009.06.031
- Ou Y, Lu X, Zi Q, Xun Q, Zhang J, Wu Y, Shi H, Wei Z, Zhao B, Zhang X, et al. RGF1 INSENSITIVE 1 to 5, a group of LRR receptor-like kinases, are essential for the perception of root meristem growth factor 1 in Arabidopsis thaliana. Cell Res 2016; 26:686-98; PMID:27229312; http://dx.doi.org/10.1038/cr.2016.63
- Wang K, Zhao Y, Li M, Gao F, Yang MK, Wang X, Li S, Yang P. Analysis of phosphoproteome in rice pistil. Proteomics 2014; 14:2319-34; PMID:25074045; http://dx.doi.org/10.1002/pmic.201400004
- Zelazny E, Barberon M, Curie C, Vert G. Ubiquitination of transporters at the forefront of plant nutrition. Plant Signal Behav 2011; 6:1597-9; PMID:21918375; http://dx.doi.org/10.4161/psb.6.10.17134
- Li J, Tax FE. Receptor‐like kinases: Key regulators of plant development and defense. J Integr Plant Biol 2013; 55:1184-7; PMID:24308569; http://dx.doi.org/10.1111/jipb.12129
- Durbak AR, Tax FE. CLAVATA signaling pathway receptors of Arabidopsis regulate cell proliferation in fruit organ formation as well as in meristems. Genetics 2011; 189: 177-94; PMID:21705761; http://dx.doi.org/10.1534/genetics.111.130930
