ABSTRACT
Protein S-acyl transferases (PATs) play critical roles in plant developmental and environmental responses by catalyzing S-acylation of substrate proteins, most of which are involved in cellular signaling. However, only few plant PATs have been functionally characterized. We recently demonstrated that Arabidopsis PAT4 mediates root hair elongation by positively regulating the membrane association of ROP2 and actin microfilament organization. Here, we show that apex-associated re-positioning of nucleus during root hair elongation was impaired by PAT4 loss-of-function. Results presented here pose a significant question concerning the molecular machinery mediating nuclear migration during root hair growth.
S-acylation, or as commonly named, palmitoylation, is a reversible post-translational modification regulating the subcellular targeting, activity, and interaction profiles of substrate proteins.Citation1-3 Plenty of proteins, especially those related to cellular signaling such as small GTPases, receptor-like cytoplasmic kinases (RLCKs), SNAREs, are subject to palmitoylation based on proteomic analyses in yeast, mammals, and plants.Citation4-6 The vast number of palmitoylated, signaling-related proteins suggests the importance of protein palmitoylation on the development and environmental responses of eukaryotes.
A class of Asp-His-His-Cys motif Cys-rich domain (DHHC-CRD) protein S-acyl transferases (PATs) is mainly responsible for the catalysis of protein palmitoylation.Citation1,2,7 PATs are present in most plant genomes as large gene families, such as 24 members in Arabidopsis and 40 members in maize.Citation8,9 Despite the potential importance of PATs in plant development and environmental responses, there are only few whose functionality has been characterized.Citation9-15 We recently reported that Arabidopsis PAT4, a member of the protein S-acyl transferase family, mediates root hair elongation.Citation16 PAT4 is expressed preferentially in tip-growing cells, i.e. root hairs and pollen tubes.Citation16 Interestingly, its expression is highest in elongating root hairs, suggesting its function during rapid cell elongation. Indeed, functional loss of PAT4 resulted in shorter root hairs whereas had little effect on root hair differentiation and initiation.Citation16
We demonstrated that PAT4 targets to the plasma membrane (PM) through BFA-sensitive vesicle trafficking and its potentially enzymatic inactive mutant, although showing the same localization pattern, could not complement the short root hair phenotype.Citation16 This result suggested that PAT4 mediates the PM targeting of substrate proteins to regulate root hair elongation. Because the small GTPase ROP2 was implicated in the regulation of root hair elongationCitation17 and was likely a palmitoylated protein,Citation18 we tested the hypothesis that PAT4 mediates the palmitoylation of ROP2 and thus affects ROP signaling during root hair elongation. Although we still have not got direct biochemical data supporting the hypothesis due to technical difficulties, several lines of evidence supported PAT4-dependent palmitoylation of ROP2 in root hairs.Citation16
First, the PM-association of ROP2 was significantly reduced in pat4 root hairs, similar to that of a ROP2 mutant in which 2 palmitoylation sites were mutated, both based on fluorescence quantification and on membrane fractionation assays.Citation16 Second, growing pat4 root hairs contained abnormal actin filaments (AFs) that were bundled and penetrated to the very apex rather than stayed behind the apical clear zone.Citation16 Dynamic AF organization in root hairs is one of the most prominent intracellular activities initiated by ROP signalingCitation17 and such the abnormal AFs distribution resembled greatly to that caused by the expression of a dominant negative ROP2.Citation17 Finally, we demonstrated that PAT4 genetically interacts with RhoGDI1/SCN1, a ROP regulator whose functional loss resulted in ectopic ROP2 distribution at the PM.Citation19
Except for AFs, dynamic organization of microtubule s (MTs) is an intracellular event involved in root hair elongation such that disruption of MT dynamics pharmacologically or genetically impaired the growth of root hairs.Citation20,21 It was also reported that ROPs mediate MT distribution through effectors to regulate the jigsaw morphology of leaf pavement cells.Citation22 To test whether the distribution of MT was affected by PAT4 loss-of-function, we introduced a Pro35S: GFP-MBD into pat4–2. GFP-MBD is a microtubule reporter by fusing the microtubule binding domain of the mammalian microtubule-associated protein 4 (MAP4) gene with the green fluorescent protein (GFP) gene.Citation23 Confocal laser scanning microscopic (CLSM) examination of GFP-MBD showed that root hairs from either wild type or pat4–2 contained longitudinal and cortical MT cables along the growth axis (). No discernible differences between the 2 genotypes were observed.
Figure 1. Distribution of microtubule and nuclei in wild-type and pat4–2 root hairs. (A-B) CLSM projections of a growing root hair from Pro35S: GFP-MBD (A) or Pro35S: GFP-MBD; pat4–2 (B). (C-D) A representative primary root of wild-type (C) or pat4–2 (D) seedlings at 4 d after germination (DAG) stained with DAPI. (E) Distance between nucleus and the apex. (F) Relative distance the nucleus traveled (distance between the nucleus and the root hair base fractionated with root hair length). Results shown in (E) and (F) are means ± standard deviation (n = 30). Asterisks indicate significant difference (t-test, P < 0.05). Bars = 10 μm for (A-B); 100 μm for (C-D).
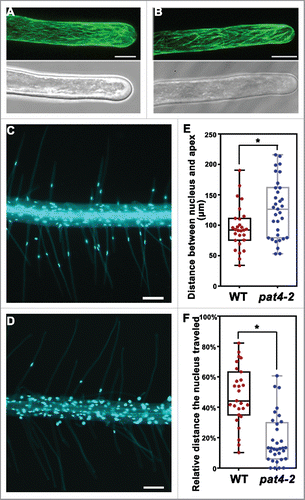
Root hair growth also accompanies with tip-associated nucleus re-localization. In Arabidopsis, the nuclei of root hairs locate at a fixed distance from the apex during growth while migrate to a random position during growth arrest.Citation24 Pharmacological studies have demonstrated that actin MF between the nucleus and the apex is required for its re-localization.Citation24 Artificially inhibiting the apex-associated nuclear migration caused growth arrest,Citation24 suggesting an important role of nuclear positioning in root hair elongation. Because pat4–2 root hairs are short and with defective AF organization,Citation16 we wondered whether the apex-associated nuclear positioning was also affected. To test this hypothesis, we stained root hairs with 4',6-diamidino-2-phenylindole (DAPI). In wild-type root hairs, nuclei migrate toward tip during root hair elongation (), as reported.Citation24 By contrast, in pat4–2 root hairs, the nucleus mostly stayed at the base of root hairs rather than coming out along with root hair growth (). Even taken the reduced length of pat4–2 root hairs into consideration, nuclear migration was much more reduced in pat4–2 than that in wild type ().
The significance of apex-associated re-localization of nucleus during root hair growth is not clear. A most obvious benefit of such positioning is to achieve local transcription to serve the needs of rapid cell elongation. Disrupting AFs abolished nuclear re-localization during root hair growthCitation24 and we showed here that functional loss of PAT4, which associates with reduced ROP signaling, affected nuclear migration. It will be interesting in the future to explore the possibility whether and how ROP-mediated AF dynamics participate in nuclear positioning during polarized cell growth in plants.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Prof. Elison B. Blancaflor for the Pro35S:GFP-MBD transgenic line. This work was supported by by Natural Science Foundation of Shandong Province (ZR2014CM027 to S. L.), and by China Postdoctoral Science Foundation funded project (2015M570605 to S. L.). The authors declare that there is no conflict of interest.
References
- Greaves J, Chamberlain LH. DHHC palmitoyl transferases: Substrate interactions and (patho)physiology. Trends Biochem Sci 2011; 36:245-53; PMID:21388813; http://dx.doi.org/10.1016/j.tibs.2011.01.003
- Hemsley PA, Grierson CS. Multiple roles for protein palmitoylation in plants. Trends Plant Sci 2008; 13:295-302; PMID:18501662; http://dx.doi.org/10.1016/j.tplants.2008.04.006
- Running MP. The role of lipid post-translational modification in plant developmental processes. Front Plant Sci 2014; 5:50; PMID:24600462; http://dx.doi.org/10.3389/fpls.2014.00050
- Hemsley PA, Weimar T, Lilley KS, Dupree P, Grierson CS. A proteomic approach identifies many novel palmitoylated proteins in Arabidopsis. New Phytol 2013; 197:805-14; PMID:23252521; http://dx.doi.org/10.1111/nph.12077
- Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R, et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 2008; 456:904-9; PMID:19092927; http://dx.doi.org/10.1038/nature07605
- Roth AF, Wan J, Bailey AO, Sun B, Kuchar JA, Green WN, Phinney BS, Yates JR 3rd, Davis NG. Global analysis of protein palmitoylation in yeast. Cell 2006; 125:1003-13; PMID:16751107; http://dx.doi.org/10.1016/j.cell.2006.03.042
- Baekkeskov S, Kanaani J. Palmitoylation cycles and regulation of protein function. Mol Membr Biol 2009; 26:42-54; PMID:19169934; http://dx.doi.org/10.1080/09687680802680108
- Batistic O. Genomics and localization of the Arabidopsis DHHC-cysteine-rich domain S-acyltransferase protein family. Plant Physiol 2012; 160:1597-612; PMID:22968831; http://dx.doi.org/10.1104/pp.112.203968
- Zhou LZ, Li S, Feng QN, Zhang YL, Zhao X, Zeng YL, Wang H, Jiang L, Zhang Y. PROTEIN S-ACYL TRANSFERASE10 is critical for development and salt tolerance in Arabidopsis. Plant Cell 2013; 25:1093-107; PMID:23482856; http://dx.doi.org/10.1105/tpc.112.108829
- Hemsley PA, Kemp AC, Grierson CS. The TIP GROWTH DEFECTIVE1 S-acyl transferase regulates plant cell growth in Arabidopsis. Plant Cell 2005; 17:2554-63; PMID:16100337; http://dx.doi.org/10.1105/tpc.105.031237
- Lai J, Yu B, Cao Z, Chen Y, Wu Q, Huang J, Yang C. Two homologous protein S-acyltransferases, PAT13 and PAT14, cooperatively regulate leaf senescence in Arabidopsis. J Exp Bot 2015; 66:6345-53; PMID:26160582; http://dx.doi.org/10.1093/jxb/erv347
- Li Y, Scott R, Doughty J, Grant M, Qi B. Protein S-acyltransferase 14: A specific role for palmitoylation in leaf senescence in Arabidopsis. Plant Physiol 2016; 170:415-28; PMID:26537563; http://dx.doi.org/10.1104/pp.15.00448
- Qi B, Doughty J, Hooley R. A Golgi and tonoplast localized S-acyl transferase is involved in cell expansion, cell division, vascular patterning and fertility in Arabidopsis. New Phytol 2013; 200:444-56; PMID:23795888; http://dx.doi.org/10.1111/nph.12385
- Zhang YL, Li E, Feng QN, Zhao XY, Ge FR, Zhang Y, Li S. Protein palmitoylation is critical for the polar growth of root hairs in Arabidopsis. BMC Plant Biol 2015; 15:50; PMID:25849075; http://dx.doi.org/10.1186/s12870-015-0441-5
- Zhao XY, Wang JG, Song SJ, Wang Q, Kang H, Zhang Y, Li S, et al. Precocious leaf senescence by functional loss of PROTEIN S-ACYL TRANSFERASE14 involves the NPR1-dependent salicylic acid signaling. Sci Rep 2016; 6:20309; PMID:26842807; http://dx.doi.org/10.1038/srep20309
- Wan ZY, Chai S, Ge FR, Feng QN, Zhang Y, Li S. Arabidopsis PROTEIN S-ACYL TRANSFERASE4 mediates root hair growth. Plant J 2017; 90:249-260; PMID:28107768; http://dx.doi.org/10.1111/tpj.13484
- Jones MA, Shen JJ, Fu Y, Li H, Yang Z, Grierson CS. The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell 2002; 14:763-76; PMID:11971133; http://dx.doi.org/10.1105/tpc.010359
- Sorek N, Gutman O, Bar E, Abu-Abied M, Feng X, Running MP, Lewinsohn E, Ori N, Sadot E, Henis YI, et al. Differential effects of prenylation and s-acylation on type I and II ROPS membrane interaction and function. Plant Physiol 2011; 155:706-20; PMID:21139084; http://dx.doi.org/10.1104/pp.110.166850
- Carol RJ, Takeda S, Linstead P, Durrant MC, Kakesova H, Derbyshire P, Drea S, Zarsky V, Dolan L. A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 2005; 438:1013-6; PMID:16355224; http://dx.doi.org/10.1038/nature04198
- Bibikova TN, Blancaflor EB, Gilroy S. Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J 1999; 17:657-65; PMID:10230063; http://dx.doi.org/10.1046/j.1365-313X.1999.00415.x
- Ketelaar T, de Ruijter NC, Emons AM. Unstable F-actin specifies the area and microtubule direction of cell expansion in Arabidopsis root hairs. Plant Cell 2003; 15:285-92; PMID:12509537; http://dx.doi.org/10.1105/tpc.007039
- Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z. Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 2005; 120:687-700; PMID:15766531; http://dx.doi.org/10.1016/j.cell.2004.12.026
- Marc J, Granger CL, Brincat J, Fisher DD, Kao Th, McCubbin AG, Cyr RJ. A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 1998; 10:1927-40. PMID:9811799; http://dx.doi.org/10.1105/tpc.10.11.1927
- Ketelaar T, Faivre-Moskalenko C, Esseling JJ, de Ruijter NC, Grierson CS, Dogterom M, Emons AM. Positioning of nuclei in Arabidopsis root hairs: an actin-regulated process of tip growth. Plant Cell 2002; 14:2941-55; PMID:12417712; http://dx.doi.org/10.1105/tpc.005892
