ABSTRACT
To gain insights into the molecular basis of starch degradation in wheat seeds with respect to dormancy maintenance and release, this study compared the expression of starch degrading genes between dormant and after-ripened seeds in both dry and imbibed states. Furthermore, the study examined the effect of ABA on the expression of starch degrading genes during imbibition of non-dormant seeds. Release of dormancy due to after-ripening led to the upregulation of specific genes encoding α-amylase and α-glucosidase during imbibition while dormancy maintenance is associated with repression of these genes. It appears from our result that ABA delays the germination of wheat seeds at least partly through repression of the starch degrading genes.
Wheat is one of the most important crops in the world; however, its yield and quality are affected by several biotic and abiotic stress factors. The occurrence of wet and humid conditions prior to harvest of cereal crops leads to preharvest sprouting (PHS), which refers to the germination of mature seed while on the mother plant, and this causes significant reduction in seed yield and quality.Citation1 The negative effects of PHS on yield and quality due to preharvest sprouting are attributed mainly to increased activities of starch degrading enzymes. Starch degradation during sprouting has been implicated in decreasing water binding capacity, swelling power and gelatinization temperature of starch, and thereby altering its physicochemical property.Citation2,Citation3 The incidence of PHS is closely associated with the level of seed dormancy, which is defined as the inability of intact viable seeds to complete germination under favorable conditions.Citation4
Degradation of storage starch is one of the metabolic processes required for the provision of energy to the growing embryos and seedlings during and after germination,Citation5 and it is mediated by the synergistic action of several amylolytic enzymes.Citation6,Citation7 α-amylase (AMY) is considered as the primary enzyme that acts on starch granules and produce dextrins.Citation8 Previous studies indicated the absence of AMY expression or its activity in mature dry seeds of cereals, however, its expression or activity has been shown to increase with germination as it is required for mobilization of storage reserves deposited in the endosperm.Citation9,Citation10,Citation11 Limit dextrinase converts the branched glucans produced by AMYs to linear glucans, and β-amylase (BAM) removes the maltose units from non-reducing ends of the linear glucans. However, BAM is considered to have less control on the production of sugars used for embryos. For example, seeds of BAM deficient barley varieties and mutant lines that exhibit low level of BAM activity can germinate and produce normal seedlings.Citation12,Citation13 The enzyme α-glucosidase (AGL) is responsible for hydrolysis of maltose to glucose during germination and early seedling growth of cereal seeds such as barley,Citation14 and a single AGL gene (designated as Agl97) has been reported to account for all the AGL activity in the endosperm of barley seedsCitation15 although four distinct barley AGL genes have been identified.Citation16
This study involved the use of dormant and after-ripened (generated by dry storage of dormant seeds at room temperature for 10 months) wheat seed samples of cv. AC Domain that exhibits a high level of dormancy/preharvest sprouting tolerance.Citation17 Germination behaviour of the seed samples was monitored using an assay described before.Citation18 The seed samples (both dormant and after-ripened) were imbibed with water for 12 and 24 h while the after-ripened seeds were also imbibed in 50 µM ABA solution for a period of 24 h. Dry seeds (imbibed for 0 h) and seeds imbibed for 12 and 24 h in water or ABA were subjected to mRNA extraction and subsequent microarray analysis as described before.Citation18 Log2 transformed signal intensities of the probesets annotated as starch degrading genes were extracted from our microarray dataset (GEO accession number GSE32409) and then converted to expression values in log2 (; Table S1) and linear scaled fold changes (Table S1). Comparison of fold changes in the expression of starch degrading genes between the seed samples was performed using cut-off values of 2-fold change (linear scaled) and P ≤ 0.05.
Figure 1. Heat maps showing changes in the expression of probesets annotated as AMY (A), BAM (B) and AGL (C) genes. Log2 scaled fold changes in the expression of probesets during imbibition of dormant (D-12/D-0, D-24/D-0 and D-24/D-12) and after-ripened seeds (AR-12/AR-0, AR-24/AR-0, AR-24/AR-12), between dormant and after-ripened seeds in both dry and imbibed states (AR-0/D-0, AR-12/D-12 and AR-24/D-24) and between water and ABA imbibed after-ripened seeds (AR-24/AR-24+ABA). Expression values in log2 fold changes are represented by the negative and positive numbers on the bar and the color scale at the top of each heat map; higher and lower expression levels of respective probesets are represented by red and green colors, respectively. Log2 and linear scaled fold changes in expression of the probesets and the respective P values can be found in Table S1.
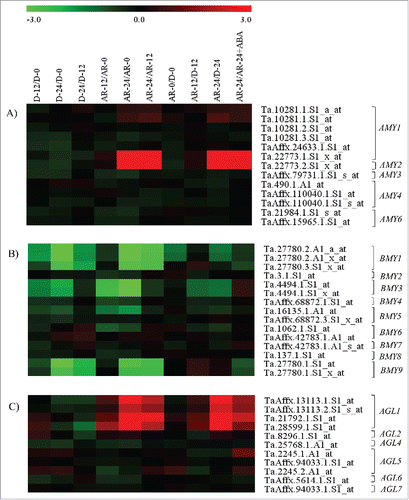
Our data indicated that 95% of the after-ripened seeds completed germination within 24 h while only 15% of the dormant seeds germinated after 5 days of imbibition.Citation19 Seed imbibition for 12 or 24 h did not affect the expression of all probesets annotated as AMY genes in the dormant seeds, however, it led to over 22-fold upregulation of two probesets each annotated as AMY1 and AMY2 in after-ripened seeds (A, Table S1), leading to their higher level of expression (over 30-fold) in after-ripened than dormant seeds. These results suggest the importance of AMY1 and AMY2 in degrading the starch deposited in wheat endosperm for supplying energy to the growing embryo axis. Previous studies also showed increases in the expression of specific AMY gene family members such as AMY1 and AMY4 in the aleurone of imbibed barley seeds.Citation20 Treatment of after-ripened seeds with ABA led to over 4-fold downregulation of probesets representing AMY1 and AMY2 genes, which exhibited imbibition mediated upregulation in the after-ripened seeds, suggesting that the role of ABA in delaying wheat seed germinationCitation21,Citation22 is mediated at least partly by suppression of starch degradation.
Seed imbibition downregulated the expression of BAM1, BAM3 and BAM9 probesets in both dormant and after-ripened seeds, although a probeset annotated as BAM5 is downregulated only in after-ripened seeds (B, Table S1). Consistently, a proteomic study revealed the downregulation of BAM isoforms during imbibition in both dormant and after-ripened seeds of wheat.Citation23 Comparative analysis of the expression of probesets annotated as BAM genes between dormant and after-ripened seeds in both dry and imbibed states revealed over 2-fold downregulation of those representing BAM1 and BAM3 in the non-dormant/after-ripened seeds, suggesting the minimal role of BAM in reserve mobilization and therefore embryo growth.Citation12,Citation13 Treatment of after-ripened seeds with ABA did not affect the expression of all probesets annotated as BAM genes, although ABA has been shown to inhibit BAM activity in rice seeds.Citation24
The expression of probesets annotated as AGL did not show any change during imbibition of dormant seeds (C, Table S1), however, probesets annotated as AGL1 exhibited upregulation (4.5- to 28-fold) in the corresponding after-ripened seeds, leading to their higher level expression (5.5- to 35-fold) in the after-ripened than the corresponding dormant seeds. These results suggest the significance of AGL1 in the hydrolysis of maltose to glucose during wheat seed germination. Treatment of after-ripened seeds with ABA led to downregulation (1.8- to 8-fold) of AGL1 probesets, implying that suppression of AGL1 contributes to the delay of germination induced by ABA.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
supp_data_1411449.xlsx
Download MS Excel (35.1 KB)Additional information
Funding
References
- Olaerts H, Roye C, Derde LJ, Sinnaeve G, Meza WR, Bodson B, Courtin CM. Impact of preharvest sprouting of wheat (Triticum aestivum) in the field on starch, protein, and arabinoxylan properties. J Agric Food Chem. 2016;64:8324–32. doi:10.1021/acs.jafc.6b03140.
- Lorenz, K., Kulp, K., Collins, F. Sprouting of cereal grains – Effects on starch characteristics. Starch / Stärke. 1981;33:183–87. doi:10.1002/star.19810330603.
- Simsek, S., Ohm, J.-B., Lu, H., Rugg, M., Berzonsky, W., Alamri, M., Mergoum, M. Effect of pre-harvest sprouting on physicochemical properties of starch in wheat. Foods. 2014;3:194–207. doi:10.3390/foods3020194.
- Gao F, Ayele BT. Functional genomics of seed dormancy in wheat: advances and prospects. Front Plant Sci. 2014;5:458. doi:10.3389/fpls.2014.00458.
- Zeeman SC, Kossmann J, Smith AM. Starch: Its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol. 2010;61:209–34. doi:10.1146/annurev-arplant-042809-112301.
- Lloyd JR, Kossmann J, Ritte G. Leaf starch degradation comes out of the shadows. Trends Plant Sci. 2005;10:130–37. doi:10.1016/j.tplants.2005.01.001.
- Streb S, Zeeman SC. Starch metabolism in Arabidopsis. Arabidopsis Book. 2012;10:e0160. doi:10.1199/tab.0160.
- Dunn G. A model for starch breakdown in higher plants. Phytochemistry. 1974;13:1341–46. doi:10.1016/0031-9422(74)80289-X.
- Perata P, Pozueta-Romero J, Akazawa T, Yamaguchi J. Effect of anoxia on starch breakdown in rice and wheat seeds. Planta. 1992;188:611–18. doi:10.1007/BF00197056.
- Bewley JD, Black M. Seeds: Physiology of development and germination. New York (USA): Plenum Press; 1994.
- Guglielminetti L, Yamaguchi J, Perata P, Alpi A. Amylolytic activities in cereal seeds under aerobic and anaerobic conditions. Plant Physiol. 1995;109:1069–76. doi:10.1104/pp.109.3.1069.
- Kihara M, Kaneko T, Ito K, Takeda K. Geographical variation of beta amylase thermostability among varieties of barley “Hordeum vulgare” and beta amylase deficiency. Plant Breed 1999;118:453–55. doi:10.1046/j.1439-0523.1999.00397.x.
- Kaneko T, Kihara M, Ito K, Takeda K. Molecular and chemical analysis of β-amylase less mutant barley in Tibet. Plant Breed. 2000;119:383–87. doi:10.1046/j.1439-0523.2000.00502.x.
- Ling A, Nanji D. CCCVI.—Studies on starch. Part I. The nature of polymerised amylose and of amylopectin. J Chem Soc Trans. 1923;123:2666–88. doi:10.1039/CT9232302666.
- Andriotis VME, Saalbach G, Waugh R, Field RA, Smith AM. The maltase involved in starch metabolism in barley endosperm is encoded by a single gene. PLoS One. 2016b;11:1–13. doi:10.1371/journal.pone.0151642.
- Tanaka Y, Duke S, Henson CA. Alpha-glucosidases from the glycoside hydrolase family 31 in germinating seeds and seedling leaves of barley. In: Proceedings of the XXIIIrd International Carbohydrate Symposium. Whistler, Canada. American Chemical Society/National Research Canada 2006; USDA publication #196842 http://www.ars.usda.gov/research/publications/publications.htm?seq_no_115=196842&pf=1.
- Townley-Smith TF, Czarnecki EM. AC domain hard red spring wheat. Canadian J Plant Sci. 2008;88:347–50. doi:10.4141/CJPS07004.
- Gao F, Jordan MC, Ayele BT. Transcriptional programs regulating seed dormancy and its release by after-ripening in common wheat (Triticum aestivum L.). Plant Biotechnol J 2012;10:465–76. doi:10.1111/j.1467-7652.2012.00682.x.
- Gao F, Rampitsch C, Chitnis VR, Humphreys GD, Jordan MC, Ayele BT. Integrated analysis of seed promoted and mRNA oxidation reveals distinct post-transcriptional features regulating dormancy in wheat (Triticum aestivum L.) Plant Biotechnol J. 2013;11:921–32. doi:10.1111/pbi.12083.
- Radchuk VV, Borisjuk L, Sreenivasulu N, Merx K, Mock H-P, Rolletschek H, Wobus U, Weschke W. Spatiotemporal profiling of starch biosynthesis and degradation in the developing barley grain. Plant Physiol. 2009;150:190–204. doi:10.1104/pp.108.133520.
- Liu A, Gao F, Kanno Y, Jordan MC, Kamiya Y, Seo M, Ayele BT. Regulation of wheat seed dormancy by after-ripening is mediated by specific transcriptional switches that induce changes in seed hormone metabolism and signaling. PLoS One. 2013;8:e56570. doi:10.1371/journal.pone.0056570.
- Chitnis VR, Gao F, Yao Z, Jordan MC, Park S, Ayele BT. After-ripening induced transcriptional changes of hormonal genes in wheat seeds: The cases of brassinosteroids, ethylene, cytokinin and salicylic acid. PLoS One. 2014;9:e87543. doi:10.1371/journal.pone.0087543.
- Park S, Rampitsch C, Humphreys GD, Ayele BT. Wheat seed proteins regulated by imbibition independent of dormancy status. Plant Signal Behav. 2013;8:10–12. doi:10.4161/psb.26601.
- Wang SM, Lue WL, Eimert K, Chen J. Phytohormone-regulated beta-amylase gene expression in rice. Plant Mol Biol. 1996;31:975–82. doi:10.1007/BF00040716.
