 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
As an endophytic fungus of Sebacinales, Piriformospora indica promotes plant growth and resistance to abiotic stress, including drought. Colonization of maize roots promoted the leaf size, root length and number of tap roots. Under drought stress, the maize seedlings profited from the presence of the fungus and performed visibly better than the uncolonized controls. To identify genes and biological processes involved in growth promotion and drought tolerance conferred by P. indica, the root transcriptome of colonized and uncolonized seedlings was analyzed 0, 6 and 12 h after drought stress (20% polyethylene glycol 6000). The number of P. indica-responsive genes increased from 464 (no stress at 0 h) to 1337 (6 h drought) and 2037 (12 h drought). Gene Ontology analyses showed that the carbon and sulfur metabolisms are major targets of the fungus. Furthermore, the growth promoting effect of P. indica is reflected by higher transcript levels for microtubule associated processes. Under drought stress, the fungus improved the oxidative potential of the roots, and stimulated genes for hormone functions, including those which respond to abscisic acid, auxin, salicylic acid and cytokinins. The comparative analyses of our study provides systematic insight into the molecular mechanism how P. indica promotes plant performance under drought stress, and presents a collection of genes which are specifically targeted by the fungus under drought stress in maize roots.
Introduction
With the impending global climate change, drought and water deficit have become the major constraints for crop production.Citation1 As one of major crops, maize suffers from drought and water deficit stress, which results in serious yield loss.Citation2 P. indica, as an endophytic fungus of Sebacinales, has been well documented to enhance plant development, production, and tolerance to abiotic stress in several plant species.Citation3,4 The multiple beneficial effects conferred by P. indica colonization show the potential to improve maize productivity under abiotic stress such as drought.
P. indica was first reported as a novel endophyte to promoting root development.Citation3-5 Later, plants colonized with P. indica were found to be promoted in biomass accumulationCitation3,Citation5,Citation6 and nutrient uptake.Citation7 In addition, P. indica also confers abiotic stress tolerance, especially under salinity and drought stress.Citation8-12 To deal with drought stress, higher plants have evolved multiple strategies at the molecular, cellular and physiological levels.Citation13 At the cellular level, compatible solutes, e.g., amino acids, amines and carbohydrates, are produced to maintain cell turgor pressure to stabilize proteins and cellular structure.Citation14-16 Furthermore, plants have developed enzymatic and non-enzymatic antioxidant defense mechanisms to avoid oxidative damage and maintain the redox homeostasis.Citation1 At the molecular level, genes related to the synthesis of osmoprotectants, detoxifying enzymes and transporters, transcription factors, protein kinases and phosphatases are induced under drought stress.Citation16,Citation17 Notably, phytohormones play pivotal roles in coordinating various signal transduction pathways during response to environmental stress.Citation18,Citation19 Abscisic acid (ABA) is involved in drought stress responses in plants and many genes respond to this stress through ABA-dependent and ABA-independent pathways.Citation17,Citation20 In addition, auxin, cytokinin, ethylene, gibberellins, jasmonic acid, and brassinosteroids contribute to drought tolerance in plants in various ways.Citation21-25
Relatively little is known about the mechanism of drought tolerance when plants are colonized with P. indica. In Arabidopsis, drought responsive genes were induced under drought stress when colonized with P. indica.Citation26 In Chinese cabbage and maize, P. indica confers drought tolerance in leaves by stimulating antioxidant enzymes and drought-related genes.Citation10,Citation27 These studies have assigned certain roles to a few genes coding for key components in the drought stress response, but a global view on genes involved in this stress and how the gene products function remain unclear. In particular, the effect of the fungus on the expression of genes in drought-stressed roots, the primary target organ of the fungus, is little investigated.
In this study, RNA-seq was exploited to discover genes in roots differentially expressed among maize seedlings colonized with or without P. indica under drought stress. We identified genes and predict metabolic pathways as potential targets of P. indica under drought stress.
Results
Morphological changes induced by P. indica under drought stress
Maize seedlings at the three-leaf stage were analyzed 15 DAI by P. indica and compared to the uncolonized control. P. indica chlamydospores were clearly lined up intercellularly in and around maize tap root (). Compared with uncolonized maize seedlings (P. indica-), plants colonized with P. indica had more and longer leaves ().
Figure 1. Phenotype differences between maize seedlings colonized and uncolonized by P. indica. (A) Typical root architecture of maize 15 days after inoculation (left) or uninoculation (B, right) with P. indica, P. indica chlamydospores on maize tap root inspected by Trypan blue staining (white arrow) at 15 DAI under microscope (Leica DM5000 B, Germany), with scale bar=20 μm. (C, D) Typical shoot and root phenotype of maize 15 days after inoculation (right) or uninoculation (left) with P. indica, with scale bar=5 cm. (E-H): Length of tap root (E), fresh weight (FW) of roots (F), dry weight (DW) of roots (G), and leaf area (H) of maize seedlings inoculated or uninoculated with P. indica under drought stress, while uninoculation and not-drought stress-exposed seedlings were used as control. All experiments were performed with at least 3 replications. Statistically significant differences are labeled with letters (p< 0.05, two-wayfactor ANOVAone way ANOVA).
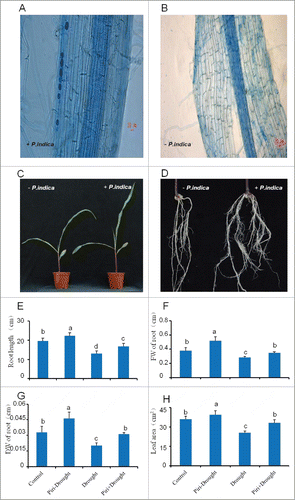
Maize seedlings co-cultivated with P. indica or mock-treated in sand for 15 days were exposed to drought for seven days, and allowed to recover for additional two days. As expected, the leaf areas of drought-exposed seedlings were smaller than those of the none-stressed controls, but in both cases, colonized plants had larger leaf areas than the uncolonized plants (). Similar results were observed for root traits (length, fresh and dry weights) (). Notably, root dry weight was significantly higher for colonized plants than for all other treatments (). Thus, the effect of P. indica on drought-exposed and control roots was investigated by pair-end sequencing based transcriptome analysis.
Pair-end sequencing based transcriptome analysis of maize tap roots infected by P. indica and under drought stress
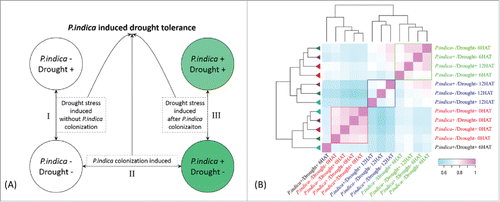
To further elucidate the mechanism of P. indica-induced drought tolerance, transcriptome analysis was carried out by RNA-seq (pair-end sequencing) for seedlings which were either colonized by P. indica for 15 days or mock-treated, before exposure to 20% polyethylene glycol (or water) for 0, 6 and 12 h.Citation28 The combination of these four treatments was repeated three times ().
Figure 2. Experiment design and expression correlation between maize genes in roots (A) Strategies to decipher P. indica-induced drought tolerance related genes. (B) Clustering of different samples by spearman correlation analysis based on FPKM (Fragment Per Kilo bases per Million reads) of each transcript. The color scale indicates the degree of correlation coefficient (blue for low correlation and pink for high correlation). Samples clustered together were indicated by rectangle box (red, blue, and green, respectively). The map was generated using R packages.
Over 720 million reads were generated from the 12 cDNA libraries. After adaptor sequences and low quality reads were removed, a total of 599 million clean reads were obtained. For each treatment, approximate 24.95 million clean reads (∼63.4% of total clean reads) were mapped to the maize genome (Zea mays cv. B73). Approximately 50.8% of the clean reads were uniquely mapped to the genome and 49.2% were mapped to multiple genomic locations. In total, ∼83.3% of genome-mapped sequences were related to annotated protein-coding gene models (www.phytozome.net/maize) (Table S1).
A total of 35 to 45 thousand transcripts mapped to the gene model were detected in each treatment set (), which accounts for an average of 63.9% of total annotated gene models in maize (B73). As outlined in , genes that showed significantly different expression levels (p<0.01, FDR<0.05) are defined as differentially expressed genes (DEGs). DEGs at each time point between different treatments spanned from approximately 2300 to 10000 [only genes with log2 fold change > 1.6 were taken into account, p<0.01, FDR<0.05] (). The gene expression pattern before PEG application (t = 0 h) was significantly different from those 6 and 12 h after the PEG treatment (6 and 12 HAT), indicating that the maize seedlings effectively responded to drought stress (, red box). Notably, changes in maize gene expression patterns between 6 HAT and 12 HAT for the different treatments were largely correlated except for 6 HAT of P. indica+/Drought+ ().
Table 1. Statistics of transcripts identified from different treatments.
Table 2. Comparison of DEGs among different treatments.
Transcripts responding to drought stress
Transcriptomic analysis identified 1337 DEGs which responded to drought stress (, Table S2), 523 (341) genes were either up- or down-regulated at both time points (6 and 12 HAT, respectively), while 473 genes were up-regulated at one and down-regulated at the other time point ().
Table 3. Statistics of DEGs and gene ontology enrichment from different treatments.
GO enrichment analysis classified these DEGs into 4 clusters, by the K-means clustering algorithm (, ). The majority of genes which are down-regulated 12 HAT code for structural constituents of the ribosomes or carboxylic acid binding proteins (Chi-square test p<0.01, FDR<0.05). Upregulated DEGs code for proteins involved in signal perception and pathogenesis. Genes were first down-and later up-regulated participate in carbohydrate metabolism, oxidoreductase activities, and in particular in phosphoric ester hydrolase and transferase activities () (Detailed GO annotation of this set of genes is shown in Table S3).
Figure 3. Classification of drought induced differentially expressed genes (DEGs) 1337 DEGs were obtained by comparing transcripts identified from P. indica-/Drought+ and P. indica-/Drought-. (I in ) (A) Average linkage clustering of drought-regulated DEGs by HCL (hierarchical clustering), where log2 (fold change) less than 1.6 were not shown. (B) Expression pattern of drought regulated DEGs by K-means clustering, where each line represents a single gene and the pink line indicates the general trend within a cluster. Significant GO terms (Chi-square test with p<0.01 and FDR<0.05) were presented for each DEGs cluster, with P and F indicating biological processes and molecular functions, respectively.
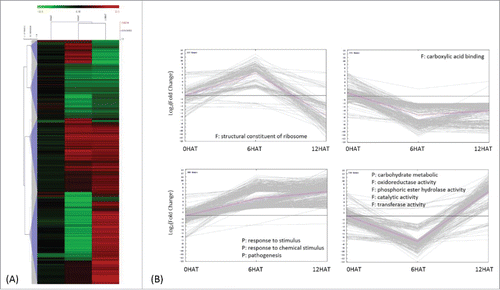
Overall, genes for carbon fixation and the C-metabolism (glyoxylate, dicarboxylate, starch, sucrose, amino acids (in particular glycine, serine, and threonine metabolism), ascorbate, pyrimidine and pyruvate metabolic pathways) were downregulated under stress, while the sulfur metabolism was up-regulated (). These results indicate that the carbon-sulfur balance has been rebalanced in response to drought stress in maize roots.
Transcripts regulated by P. indica colonization
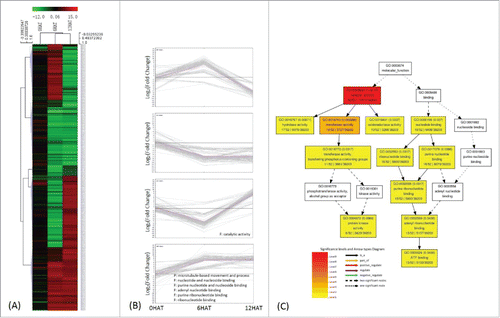
464 genes responded to P. indica in maize without stress of which 113 were up- and 132 down-regulated at both time points (6HAT and 12HAT), while 219 DEGs were up-regulated at one time point and down-regulated at the other time point (, , Table S4). GO and KEGG analyses showed that P. indica stimulates microtubule-based movements which includes genes for a β-6 tubulin (GRMZM2G172932_T02) and three kinesins (GRMZM2G033785_T01/GRMZM2G113652_T03/GRMZM2G082384_T04) (, ; Table S5). Furthermore, genes for carbon fixation, sugar metabolism, ATP synthesis and ribosomes were stimulated by the fungus, suggesting that pathways which required these components are targets of P. indica to stimulate growth of maize roots.
Figure 4. Classification of genes regulated by P. indica colonization. 464 DEGs were identified by comparing transcripts from P. indica+/Drought- and P. indica-/Drought-. (II in ) (A), (B) same as . (C) Deposition of catalytic activity (B) related genes by GO enrichment analysis, and the graph was automatically generated by agriGO.
P. indica colonization enhanced expression of drought tolerance related genes
Since P. indica-colonized maize seedlings showed enhanced drought tolerance (), the gene expression profiles were also analysed for colonized and uncolonized plants exposed to drought stress. Compared to P. indica-colonized plants not exposed to drought, 2037 genes were differentially expressed in P. indica colonized plant exposed to drought stress (; Table S7). Upregulated genes were significantly enriched for 17 biological processes, including processes involved in signal perception, stress responses, redox regulation, transport and distribution of proteins within different cellular compartments (, ; Table S5). Many of these DEGs code for membrane-localized proteins (, Table S8). Down-regulated DEGs mostly code for protein with FMN/FAD binding and oxidoreductase activities (, ). The phenylalanine, cysteine, methionine, and tryptophan metabolisms were up-regulated as well, while the nitrogen metabolism was down-regulated. Additionally, up-regulation of genes for cutin, suberine and wax biosynthesis (), suggests that these biosynthetic pathways were specific target of P. indica under drought stress. Interestingly, irrespective of the presence or absence of the fungus, genes involved in the carbon fixation, as well as in the glyoxylate and dicarboxylate metabolisms were down-regulated, while those involved in glycerophospholipid metabolism were up-regulated under drought stress ().
Figure 5. Classification of drought-regulated genes in P. indica colonized maize seedlings 2037 DEGs regulated by drought were identified by comparing transcripts from P. indica+/Drought+ and P. indica+/Drought- (III in ). (A) and (B) same as . (C) Detailed GO classification of cluster 3 in (B). Top ten terms in biological processes and molecular functions were shown, respecctively.
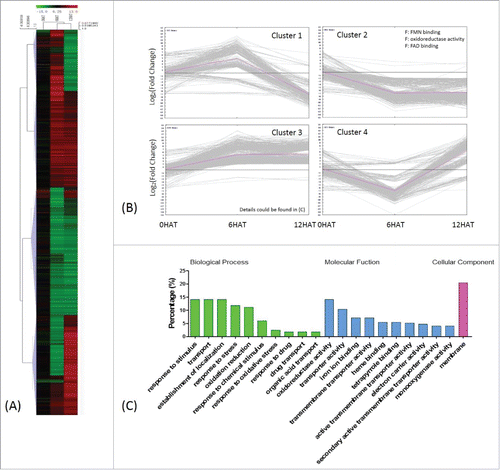
Figure 6. KEGG pathway enrichment and schematic diagram of hormone-related genes involved in drought stress tolerance. (A) DEGs detected under different conditions were analyzed by KEGG mapper, and enrichment was performed by hypergeometric probability distribution. Pathways with p<0.01 and FDR<0.05 are shown. 1 and 2: pathways commonly involved in drought stress response and drought tolerance after P. indica colonization; 3-6: pathways specific to DEG sets as denoted in the figure. (B) Schematic diagram of hormone–related genes involved in drought stress tolerance based on KEGG mapping of hormone-related DEGs, relative knowledge documented in reference (detailed annotation and expression pattern of each gene is found in the supplemental tables).
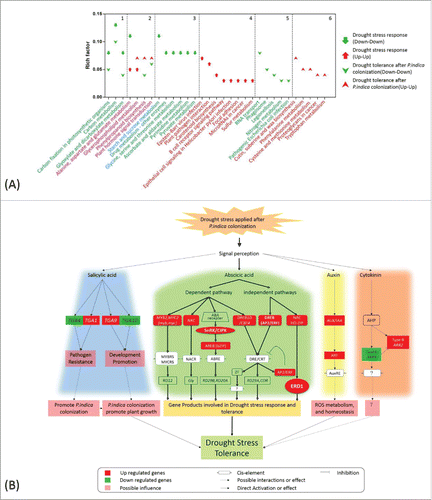
Phytohormone-related genes were involved in drought stress tolerance
Drought stress resulted in up-regulation of auxin responsive genes (Aux/IAA), genes for auxin-related transcription factors (ARF) and glutamine S-transferases (GSTs),Citation29 suggesting a positive role of auxin in the response to drought. ARR9 (GRMZM2G319187_T03), a type A gene of the cytokinin signaling pathway, was inhibited when drought stress was applied to P. indica-colonized maize seedlings, while the type B gene ARR2 (GRMZM2G126834_T02) was induced (). SnRK2s is a key component activated by osmotic stress that plays a significant role in ABA signaling.Citation30 In our study, different homologues of SnRK2.6 (GRMZM2G138861_T01) were either up- or down-regulation in P. indica uncolonized seedlings under drought stress (). This holds also true for MYB, NAC, and AP2/ERF genes involved in ABA signaling (). In addition, the gene for the salicylic acid inducible basic leucine zipper transcription factor TGA1/4 (GRMZM2G131961_T01/GRMZM2G131961_T04) was down-regulated (), while TGA9/10 (GRMZM2G030877_T01/GRMZM2G006578_T03) exhibited a disparate expression pattern in response to drought stress (). These data demonstrate that the fungus targets phytohormone-dependent signaling pathways to counteract drought stress in maize roots.
Discussion
P. indica-stimulated maize growth involves microtubule process
After 15 days, P. indica had successfully colonized the roots of maize seedlings. At this stage, the fungus should target genes for growth promotion and to a lesser extend those involved in early phases of root recognition and/or colonization. We observed the stimulation of genes involved in microtubule-based movement processes. Microtubules, together with actin microfilaments, play central roles in plant growth and development, by activating cell division, cell polarity, cell wall deposition, intracellular trafficking and communication.Citation31,Citation32 In particular, we observed a strong response of genes for a tubulin and three kinesins to P. indica (GO: 0007017, ; Table S5). Tubulin contributes to microtubule formation, and kinesin is reported to function as a molecular motor for directional transport of cellular cargo along microtubule tracks.Citation31,Citation33 In addition, microtubules also serve as sensors for mechanical membrane stress, as well as osmotic and cold stress,Citation34 suggesting that they are important targets of P. indica to promote growth of maize roots. Growth stimulating genes and compounds involved in the response to P. indica colonization have been reported repeatedly, however a dominant role of microtubules appears to be a new feature in the response of maize roots to the fungus.
Reprogramming of the carbon and sulfur metabolism under drought stress
Drought stress results from water deficit in soil, which consequently impedes plant growth.Citation35 The leaf area, root length and root fresh weight were higher in maize seedlings colonized with P. indica compared to the uncolonized control plants under drought stress (). Apparently, under our stress conditions, the plant can still coupe with the drought stress and utilizes resources for the promotion of growth when the fungus is present.
While plants reduce their investments in growth and general maintenance under drought stress, components for signal perception and protectant increase ().Citation36 Up-regulation of DEGs involved in carbohydrate metabolism along with those for oxidoreductase and catabolic activities, as well as phosphoric ester hydrolase and transferase activities (B) indicates that the plant counteracts oxidative stress by mobilizing carbohydrates to protect from injury. Since we observed a simultaneous reduction of the transcript levels for proteins involved in plastid functions under drought stress (), we propose that the plant utilizes its available C resources to respond to the drought stress (cf. also Pinheiro and Chaves).Citation37 Although several other studies obtained comparable results, the authors came to different conclusions. Sadok et al.Citation38 proposed for maize that leaf elongation under water deficit is uncoupled from the carbon status. Tardieu et al.Citation39 and Hummel et al.Citation40 hypothesized for Arabidopsis that an increase in C surplus during early phases of drought stress is caused by growth inhibition but the C is immediately utilized to maintain basic processes in the cells upon drought stress (cf. also McDowell).Citation41 Therefore, understanding of how plants reprogram their C metabolism under drought stress requires further investigations.
Notably, the sulfur metabolism was induced in the roots of drought stressed maize seedlings (A). Sulfate is the only xylem-borne metabolite regulating ABA-induced closure of stomata in leaves of maize.Citation36,Citation42 Also other drought-related processes require S. Cysteine and methionine are sulfur containing amino acids, and required for the synthesis of glutamine,Citation43 which adjusts the osmotic status of cells under drought stress. S is also a precursor for the synthesis of glutathione, an osmoprotectant functioning as a scavenger of ROS.Citation44,Citation45
Collectively, reprogramming of the carbon and sulfur metabolisms coordinates drought stress responses in maize roots.
Different phytohormones work concertedly to accommodate drought tolerance after P. indica colonization
Besides ABA, cytokinin, auxin, ethylene, gibberellins, jasmonic acid and brassinosteroids play crucial roles in various drought responses in many plant species.Citation17,Citation21-23,Citation25 Key components of the ABA, auxin, cytokinin and salicylic acid signaling pathways were identified in our study, and they are specifically upregulated in P. indica-colonized roots mediating them targets of the fungus in the drought tolerance response. Salicylic acid is a hormone mediator usually required for resistance against virulent and avirulent pathogens.Citation46 TGA1/4 is a negative regulator of pathogen resistance, and its down-regulation positively affects P. indica colonization by overcoming the plant protection systems. The response of two additional TGA genes for the TGA members 9 and 10 of the basic leucine zipper transcription factor family indicate the participation of salicylic acid in the P. indica-conferred drought tolerance response. Cytokinin is involved in drought stress response,Citation47 and our study demonstrates the involvement of both type A (ARR9) and type B (ARR2) genes in P. indica during drought. Taken together, different pathways activated by different phytohormones are targets of P. indica to concertedly accommodate drought tolerance in maize roots.
Conclusions
P. indica promotes growth and confers drought tolerance in maize. Transcriptome analysis identified DEGs which specifically responded to P. indica colonization, drought response, and drought tolerance after P. indica colonization, respectively. GO and KEGG analyses showed that P. indica might promote growth of maize roots by stimulation microtubular processes and confer drought tolerance through strengthened its redox capacity, by rebalancing the carbon-sulfur surplus and by activating hormone mediated signaling pathways.
Materials and methods
Plant materials, fungal culture and growth conditions
Seeds of maize (Zea mays cv. Jixiang 1) were surface sterilized and germinated on tissue paper at room temperature. Seedlings were transplanted into sterilized vermiculite-filled or sand-filled pots and inoculated with 50 ml P. indica (by OD600=0.08) in PNM medium. Sterilized P. indica broth was used as mock-inoculated control. Maize plants were irrigated with modified Hoagland nutrient solution every two daysCitation10 and kept in a growth chamber under conditions of 28℃, 16 h/25℃, 8/16 h day-night with a light intensity of 700 μmol·m−2·s−1. P. indica was cultured according to Rai et al.Citation48 The colonization status of P. indica on tap roots was checked under a microscope (Leica DM5000 B, Germany) 15 DAI by Trypan blue staining as described in a previous work.Citation49
Drought stress treatment
To study the physiological growth difference and explore genes responsible for drought stress in plants either inoculated with P. indica or not, drought stress was performed both under natural growth conditions and mock conditions 15 DAI. For plants grown in sand under natural conditions, irrigation was ceased after seven days and resumed for two days. After treatment, plants were harvested for measurement of leaf area, root length, root fresh weight and root dry weight. Plants grown in vermiculite were first washed with running water and then treated with 20% PEG6000 in Hoagland nutrient solution to mimic drought stress for time points of 0 h, 6 h and 12 h in a glass beaker.Citation28 Modified Hoagland nutrient solution alone was used as a control. Roots harvested for each sample were quickly frozen in liquid nitrogen and stored at -80℃ for further experiments.
RNA isolation, library construction and transcriptomic analysis
Total RNA of root tissue was extracted using TRIzol (Invitrogen) according to the user's manual. One microgram of total RNA was used to construct a pair-end index library. First and second strand cDNA was synthesized using ProtoScript II Reverse Transcriptase and Second Strand Synthesis Enzyme Mix, respectively. Then, the cDNA was purified using AxyPrep Mag PCR Clean-up (Axygen), followed by end repairing, 5’ phosphorylation, dA-tailing and adaptor ligation. Fragments of ∼400 bp (with the approximate insert size of 250 bp) were recovered and amplified by PCR (11 cycles) using P5 and P7 primers. The PCR products were cleaned up, validated and quantified using AxyPrep Mag PCR Clean-up (Axygen), an Agilent 2100 Bioanalyzer and Qubit and real-time PCR (Applied Biosystems) kits, respectively. Then, the libraries were sequenced by an Illumina HiSeq (Illumina, San Diego, CA, USA). The raw sequences were processed and analyzed by GENEWIZ using the NGSQC Toolkit (v2.3).
Analysis of differential expressed gene
The relative transcript abundance was output as FPKM (Fragment Per Kilo bases per Million reads), which is calculated as FPKM=109C/NL, where C is the number of reads mappable to the specific unigene, N is the total reads mapped to all unigenes in a certain sample, and L represents the length of the unigene. Differentially expressed genes between samples were identified by packages (edgeR) of Bioconductor with default parameters. Genes with absolute value of log2FC (fold change) larger than 1, p<0.01 (Chi-square test) and FDR (false discovery rate) <0.05 were regarded as differentially expressed. Significantly expressed genes between samples were defined as differentially expressed at 6 and 12 HAT, and genes at 0 HAT were subtracted to eliminate background noise. Perl scripts were manually developed to handle sequence extraction and dataset format.
Complete gene ontology (GO) annotation of the differentially expressed genes was performed by Singular Enrichment Analysis in a web based GO analysis tool (AgriGO, http://bioinfo.cau.edu.cn/agriGO/analysis.php)Citation50 with a Chi-square test cutoff value of p<0.01 and FDR<0.05 (Hochberg).
KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis was performed by KEGG mapper (http://www.genome.jp/kegg/mapper.html). Genes participating in specific pathways were enriched by hypergeometric probability distribution, which is calculated byWhere N is the number of all genes with pathway annotation, n refers to the number of differentially expressed genes, M is the number of genes annotated that are involved in a specific pathway, and m is the number of differentially expressed genes involved in the specific pathway. Only pathways with p<0.01 and FDR <0.05 were used for further analysis.
Abbreviations
| ABA | = | abscisic acid |
| DEG | = | differentially expressed gene |
| FC | = | fold change |
| FPKM | = | Fragment Per Kilo bases per Million reads |
| FDR | = | false discovery rate |
| GST | = | glutamine S-transferase |
| GO | = | gene ontology |
| HAT | = | hours after treatment |
| KEGG | = | Kyoto Encyclopedia of Genes and Genomes |
| P. indica | = | Piriformospora indica |
| PEG | = | polyethylene glycol |
| ROS | = | reactive oxygen species |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Suppl_Tables_Drought_stress_responses_in_maize_are_diminished_by_Piriformospora_indica.xls
Download MS Excel (1.6 MB)Additional information
Funding
References
- Fang Y, Xiong L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell Mol Life Sci: CMLS. 2015;72:673–89. doi:10.1007/s00018-014-1767-0.
- Lopes MS, Araus JL, van Heerden PD, Foyer CH. Enhancing drought tolerance in C(4) crops. J Exp Bot. 2011;62:3135–53. doi:10.1093/jxb/err105.
- Sahay NS, Varma A. Piriformospora indica: a new biological hardening tool for micropropagated plants. FEMS Microbiol Lett. 1999;181:297–302 doi:10.1111/j.1574-6968.1999.tb08858.x.
- Lou B, Sun C, Cai D. Piriformaspora indica with multiple functions and its application prospects. Acta Phytophylacica Sinica. 2007;34:653–6
- Nautiyal C, Chauhan P, DasGupta S, Seem K, Varma A, Staddon W. Tripartite interactions among Paenibacillus lentimorbus NRRL B-30488, Piriformospora indica DSM 11827, and Cicer arietinum L. J Microbiol Biotech. 2010;26:1393–99. doi:10.1007/s11274-010-0312-z.
- Anith KN, Faseela KM, Archana PA, Prathapan KD. Compatibility of Piriformospora indica and Trichoderma harzianum as dual inoculants in black pepper (Piper nigrum L.). Symbiosis. 2011;55:11–7. doi:10.1007/s13199-011-0143-1.
- Yadav V, Kumar M, Deep DK, Kumar H, Sharma R, Tripathi T, Tuteja N, Saxena AK, Johri AK. A phosphate transporter from the root endophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. J Biol Chem. 2010;285:26532–44. doi:10.1074/jbc.M110.111021.
- Baltruschat H, Fodor J, Harrach BD, Niemczyk E, Barna B, Gullner G, Janeczko A, Kogel KH, Schafer P, Schwarczinger I, Zuccaro A, Skoczowski A. Salt tolerance of barley induced by the root endophyte Piriformospora indica is associated with a strong increase in antioxidants. The New Phytologist. 2008;180:501–10. doi:10.1111/j.1469-8137.2008.02583.x.
- Waller F, Achatz B, Baltruschat H, Fodor J, Becker K, Fischer M, Heier T, Huckelhoven R, Neumann C, von Wettstein D, Franken P, Kogel KH. The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc Natl Acad Sci USA. 2005;102:13386–91. doi:10.1073/pnas.0504423102.
- Waller F, Achatz B, Baltruschat H, Fodor J, Becker K, Fischer M, Heier T, Huckelhoven R, Neumann C, von Wettstein D, Franken P, Kogel KH. The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc Natl Acad Sci USA. 2005;102:13386–91. doi:10.1073/pnas.0504423102.
- Sun C, Johnson J M, Cai D, Sherameti I, Oelmüller R, Lou B. Piriformospora indica confers drought tolerance in Chinese cabbage leaves by stimulating antioxidant enzymes, the expression of drought-related genes and the plastid-localized CAS protein. Plant Physiol. 2010;167:1009–17. doi:10.1016/j.jplph.2010.02.013.
- Jogawat A, Saha S, Bakshi M, Dayaman V, Kumar M, Dua M, Varma A, Oelmüller R, Tuteja N, Johri AK. Piriformospora indica rescues growth diminution of rice seedlings during high salt stress. Plant Signal Behav. 2013;8(10):e2689. doi:10.4161/psb.26891.
- Fathi A, Tari D B. Effect of drought stress and its mechanism in plants. Int J Life Sci. 2016;10(1):1. doi:10.3126/ijls.v10i1.14509.
- Bartels D, Sunkar R. Drought and salt tolerance in plants. Crit Rev Plant Sci. 2005;24:23–58 doi:10.1080/07352680590910410.
- Valliyodan B, Nguyen HAT. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr Opin Plant Biol. 2006;9:189–95. doi:10.1016/j.pbi.2006.01.019.
- Krasensky J, Jonak C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot. 2012;63:1593–608. doi:10.1093/jxb/err460.
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2007;58:221–7. doi:10.1093/jxb/erl164.
- Tuteja N, Sopory SK. Chemical signaling under abiotic stress environment in plants. Plant Signal Behav. 2008;3:525–36 doi:10.4161/psb.3.8.6186.
- Wolters H, Jurgens G. Survival of the flexible: hormonal growth control and adaptation in plant development. Nat Rev Genet. 2009;10:305–17. doi:10.1038/nrg2558.
- Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10:88–94. doi:10.1016/j.tplants.2004.12.012.
- de Ollas C, Hernando B, Arbona V, Gomez-Cadenas A. Jasmonic acid transient accumulation is needed for abscisic acid increase in citrus roots under drought stress conditions. Physiol Plantarum. 2013;147:296–306. doi:10.1111/j.1399-3054.2012.01659.x.
- Liu F, Xing S, Ma H, Du Z, Ma B, Cytokinin-producing, plant growth-promoting rhizobacteria that confer resistance to drought stress in Platycladus orientalis container seedlings. Appl Microbiol Biotech. 2013;97:9155–64. doi:10.1007/s00253-013-5193-2.
- Farooq M, Hussain M, Wahid A, Siddique KHM. Drought stress in plants: an overview. In: Aroca R (ed) Plant responses to drought stress. Springer Berlin Heidelberg, 2012; pp 1–33. doi:10.1007/978-3-642-32653-0_1
- Shi H, Chen L, Ye T, Liu X, Ding K, Chan Z. Modulation of auxin content in Arabidopsis confers improved drought stress resistance. Plant Physiol and Biochem: PPB / Societe Francaise de Physiologie Vegetale 2014;82:209–17. doi:10.1016/j.plaphy.2014.06.008 doi:10.1016/j.plaphy.2014.06.008.
- Tamiru M, Undan JR, Takagi H, Abe A, Yoshida K, Undan JQ, Natsume S, Uemura A, Saitoh H, Matsumura H, Urasaki N, Yokota T, Terauchi R. A cytochrome P450, OsDSS1, is involved in growth and drought stress responses in rice (Oryza sativa L.). Plant Mol Biol. 2015;88:85–99. doi:10.1007/s11103-015-0310-5.
- Sherameti I, Tripathi S, Varma A, Oelmuller R. The root-colonizing endophyte Pirifomospora indica confers drought tolerance in Arabidopsis by stimulating the expression of drought stress-related genes in leaves. Mol Plant-Microbe Interact: MPMI. 2008a;21:799–807. doi:10.1094/MPMI-21-6-0799.
- Xu L, Wang A, Wang J, et al. Piriformospora indica confers drought tolerance on Zea mays L.through enhancement of antioxidant activity and expression of drought-related genes. Crop J. 2017;5:251–58. doi:10.1016/j.cj.2016.10.002.
- Zheng J, Zhao J, Tao Y, Wang J, Liu Y, Fu J, Jin Y, Gao P, Zhang J, Bai Y, Wang G. Isolation and analysis of water stress induced genes in maize seedlings by subtractive PCR and cDNA macroarray. Plant Mol Biol. 2004;55:807–23. doi:10.1007/s11103-005-1969-9.
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell. 2001;13:2809–22. doi:10.1105/tpc.13.12.2809.
- Boudsocq M, Barbier-Brygoo H, Lauriere C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem. 2004;279:41758–66. doi:10.1074/jbc.M405259200 doi:10.1074/jbc.M405259200.
- Breviario D, Gianì S, Morello L. Multiple tubulins: evolutionary aspects and biological implications. Plant Journal. 2013;75:202–18. doi:10.1111/tpj.12243.
- Komis G, Luptovčiak I, Doskočilová A, Šamaj J. Biotechnological aspects of cytoskeletal regulation in plants. Biotech Advances. 2015;33:1043–1062. doi:10.1016/j.biotechadv.2015.03.008.
- Ganguly A, Dixit R. Mechanisms for regulation of plant kinesins. Curr Opin Plant Biol. 2013;16:704–9. doi:10.1016/j.pbi.2013.09.003.
- Nick P. Microtubules, signalling and abiotic stress. Plant J. 2013;75:309–23. doi:10.1111/tpj.12102.
- Wallace JG, Zhang X, Beyene Y, Semagn K, Olsen M, Prasanna BM, Buckler ES. Genome-wide association for plant height and flowering time across 15 tropical maize populations under managed drought stress and well-watered conditions in Sub-Saharan Africa. Crop Sci. 2016;56:1–14. doi:10.2135/cropsci2015.10.0632.
- Chan KX, Wirtz M, Phua SY, Estavillo GM, Pogson BJ. Balancing metabolites in drought: the sulfur assimilation conundrum. Trends Plant Sci. 2013;18:18–29. doi:10.1016/j.tplants.2012.07.005.
- Pinheiro C, Chaves MM. Photosynthesis and drought: can we make metabolic connections from available data? J Exp Bot. 2011;62 (3):869–82. doi:10.1093/jxb/erq340.
- Sadok W, Naudin P, Boussuge B, Muller B, Welcker C, Tardieu F. Leaf growth rate per unit thermal time follows QTL-dependent daily patterns in hundreds of maize lines under naturally fluctuating conditions. Plant Cell Environ. 2007;30:135–46. doi:10.1111/j.1365-3040.2006.01611.x.
- Tardieu F, Reymond M, Hamard P, Granier C, Muller B. Spatial distributions of expansion rate, cell division rate and cell size in maize leaves: a synthesis of the effects of soil water status, evaporative demand and temperature. J Exp Bot. 2000;51:1505–14. doi:10.1093/jexbot/51.350.1505.
- Hummel I, Pantin F, Sulpice R, Piques M, Rolland G, Dauzat M, Christophe A, Pervent M, Bouteille M, Stitt M, Gibon Y, Muller B. Arabidopsis plants acclimate to water deficit at low cost through changes of carbon usage: an integrated perspective using growth, metabolite, enzyme, and gene expression analysis. Plant Physiol. 2010;154:357–72. doi:10.1104/pp.110.157008.
- McDowell NG. Mechanisms linking drought, hydraulics, carbon metabolism, and vegetation mortality. Plant Physiol. 2011;155:1051–59. doi:10.1104/pp.110.170704.
- Ernst L, Goodger JQ, Alvarez S, Marsh EL, Berla B, Lockhart E, Jung J, Li P, Bohnert HJ, Schachtman DP. Sulphate as a xylem-borne chemical signal precedes the expression of ABA biosynthetic genes in maize roots. J Exp Bot. 2010;61:3395–405. doi:10.1093/jxb/erq160.
- Garber AJ, Karl IE, Kipnis DM. Alanine and glutamine synthesis and release from skeletal muscle. II. The precursor role of amino acids in alanine and glutamine synthesis. J Biol Chem. 1976;251:836–43
- Hasanuzzaman M, Nahar K, Anee TI, Fujita M. Glutathione in plants: biosynthesis and physiological role in environmental stress tolerance. Physiol Mol Biol Plants. 2017:23;249. doi:10.1007/s12298-017-0422-2.
- Foyer CH, Noctor G. Ascorbate and glutathione: the heart of the redox hub. Plant Physiology. 2011, 155:2–18. doi:10.1104/pp.110.167569.
- Shearer HL, Cheng YT, Wang L, Liu J, Boyle P, Despres C, Zhang Y, Li X, Fobert PR. Arabidopsis clade I TGA transcription factors regulate plant defenses in an NPR1-independent fashion. Mol Plant-Microbe Interact MPMI. 2012;25:1459–68. doi:10.1094/MPMI-09-11-0256.
- Ohri P, Bhardwaj R, Bali S, Kaur R, Jasrotia S, Khajuria A, Parihar RD. The common molecular players in plant hormone crosstalk and signaling. Curr Protein Pept Sci. 2015;16:369–88. doi:10.2174/1389203716666150330141922.
- Rai M, Acharya D, Singh A, Varma A. Positive growth responses of the medicinal plants Spilanthes calva and Withania somnifera to inoculation by Piriformospora indica in a field trial. Mycorrhiza. 2001;11:123–8. doi:10.1007/s005720100115.
- Kumar M, Yadav V, Tuteja N, Johri AK. Antioxidant enzyme activities in maize plants colonized with Piriformospora indica. Microbiology. 2009;155:780–90. doi:10.1099/mic.0.019869-0.
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–70. doi:10.1093/nar/gkq310.
