ABSTRACT
The phytohormone jasmonates (JAs) regulate plant development, growth, secondary metabolism, and defense responses. JAs act through CORONATINE INSENSITIVE1 (COI1) to induce the degradation of JA ZIM-domain (JAZ) proteins, and activate JAZ-repressed transcription factors to regulate plant response. We previously showed that the basic helix-loop-helix (bHLH) and MYB members of the WD-repeat/bHLH/MYB complex interacted with JAZs and mediated JA-induced anthocyanin accumulation and trichome initiation. In this study, we showed that the C-terminal domain of the bHLH members (GLABRA3 [GL3], ENHANCER OF GLABRA3 [EGL3] and TRANSPARENT TESTA8 [TT8]) interacted with JAZs in yeast and plant, and mediated dimerizations between the bHLH members. Our study provides further understanding of the bHLH members of the WD-repeat/bHLH/MYB complex in JA pathway.
Jasmonates (JAs), a class of cyclic fatty acid-derived phytohormones,Citation1,Citation2 regulate plant developmental processes, including plant fertility, root growth, trichome formation and senescence,Citation3-5 control secondary metabolism,Citation6,Citation7 and mediate plant defense responses against insects attack and pathogen invasion.Citation8-10 JA signals are perceived by the F-box protein CORONATINE INSENSITIVE1 (COI1), which recognizes and ubiquitinates JA ZIM-domain (JAZ) proteins for degradation, and it will release multiple JAZ-inhibited transcription factors to activate various JA response.Citation1,Citation11-16
In Arabidopsis, the WD-repeat/bHLH/MYB complex, consisting of the WD-repeat protein TRANSPARENT TESTA GLABRA1 (TTG1), the bHLH transcription factors (GLABRA3 [GL3], ENHANCER OF GLABRA3 [EGL3] and TRANSPARENT TESTA8 [TT8]), and the R2R3-MYB transcription factors (MYB75, MYB90, MYB113, MYB114, GLABRA1 [GL1]), activates expression of anthocyanin late biosynthetic genes and trichome developmental genes to promote anthocyanin accumulation and trichome initiation.Citation17-19 JAZs interact with bHLH, and MYB members of the WD-repeat/bHLH/MYB complex to inhibit their transcriptional activity, and JAs induce JAZs degradation to activate the WD-repeat/bHLH/MYB complex and enhance anthocyanin accumulation and trichome formation.Citation20
In this study, we further mapped the domain responsible for the interactions between bHLH members (GL3/EGL3/TT8) and JAZs. EGL3, GL3 and TT8 contain a JAZ-interaction domain (JID) at N-terminus,Citation21 a bHLH domainCitation22 and a conserved C-terminal domain (CD) at C-terminus (, Fig. S1). EGL3 and TT8 were respectively divided into EGL3NT and TT8NT containing the JID domain, and EGL3CT and TT8CT with bHLH and CD domains (, ). Yeast-two hybrid (Y2H) assay showed that JAZ2 and JAZ11 interacted with EGL3CT and TT8CT, but not EGL3NT and TT8NT (, ), implying that bHLH and/or CD domains are essential for interaction with JAZs, while JID is not. GL3 was truncated into GL3NT with JID and bHLH domains, and GL3CT containing bHLH and CD domains (). Y2H analysis showed that JAZ2 and JAZ11 interacted with GL3CT, but not GL3NT (), suggesting that CD domain is responsible for interaction with JAZs. Moreover, JAZ1 and JAZ8 also interacted with GL3CT, but not GL3NT ().
Figure 1. The CD domains of EGL3, TT8 and GL3 are responsible for interactions with JAZ proteins and dimeric interactions. (A) to (C) Schematic structures of EGL3, TT8 and GL3. JID, JAZ-interaction domain; CD, C-terminal domain. (D) to (H) Y2H assay to detect interactions of the truncated domains of EGL3, TT8 and GL3 with JAZ1, JAZ2, JAZ8 or JAZ11. JAZs, TTG1 and the domains of GL3, EGL3 and TT8 were fused with LexA DNA binding domain (BD) or activation domain (AD) respectively. (I) to (K) BD-fused EGL3 and GL3CT2 interact with AD-fused CT and CT2 of GL3, EGL3 and TT8 in yeast. All the interactions were detected on SD/Gal/Raf/X-gal (-Ura/-His/-Trp/-Leu) medium.
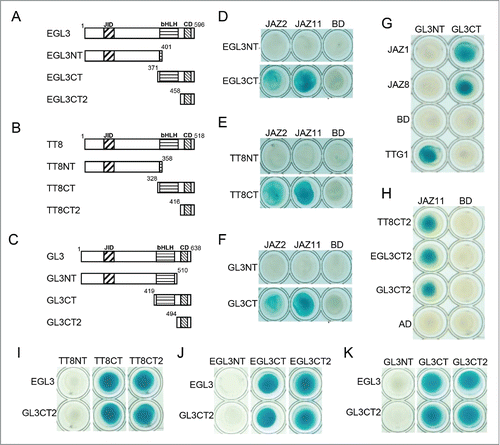
To further verify whether CD domain is responsible for interactions with JAZs. GL3, EGL3 and TT8 were further truncated into GL3CT2, EGL3CT2 and TT8CT2 harboring the CD domain (). The results showed that all the GL3CT2, EGL3CT2 and TT8CT2 interacted with JAZ11 in yeast, suggesting that the CD domain at CT2 is responsible for interaction with JAZs ().
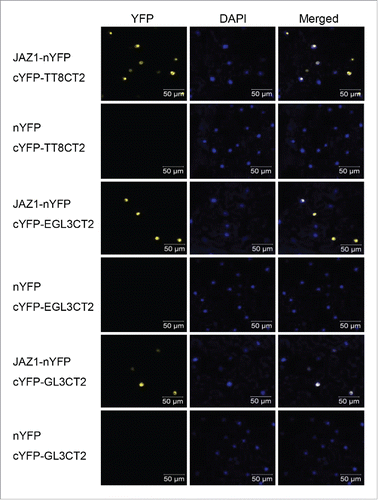
We further employed a yellow fluorescence protein (YFP) based bimolecular fluorescence complementation (BiFC) assayCitation23 to verify the interactions of CD domain with JAZ1. As shown in , coexpression of cYFP-fused GL3CT2, EGL3CT2, or TT8CT2 with nYFP-fused JAZ1 produced YFP fluorescence in the nuclei, while the negative controls did not, suggesting that CD domain of GL3, EGL3 and TT8 interacts with JAZ1 in plant. Taken together, these data demonstrated that GL3, EGL3 and TT8 interacted with JAZs through the CD domain ( & ).
Figure 2. The CT2 domains of GL3, EGL3 and TT8 interact with JAZ1 in plant. GL3CT2, EGL3CT2 and TT8CT2 were fused with C-terminal fragment of YFP (cYFP), and JAZ1 was fused with N-terminal fragment of YFP (nYFP). Agrobacterium strains containing indicated constructs pairs were co-infiltrated into the N. benthamiana leaves. The nuclei were stained by DAPI, and YFP fluorescence was tested 50h after co-infiltration.
GL3 was shown to form dimers.Citation24 We next examined the dimerization of derivatives of GL3, EGL3 and TT8 in yeast. Both EGL3 and GL3CT2 exhibited interactions with TT8CT, TT8CT2, EGL3CT, EGL3CT2, GL3CT and GL3CT2, but not TT8NT, EGL3NT and GL3NT (–), demonstrating that CD domain, but not the bHLH domain, mediates the dimerization of GL3, EGL3 and TT8.
Previous studies showed that the JID domain of the IIIe bHLH factors (e.g. MYC2) mediated interactions with JAZs.Citation16,Citation21 In this study, our results suggested that the CD domain at C-terminus of GL3, EGL3 and TT8 was essential for both interactions with JAZs and their dimerization, providing new perspectives on the interactions of JAZs with bHLH factors, and dimerization of bHLH factors.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
supp_data_1422460.tif
Download TIFF Image (24.4 MB)Additional information
Funding
References
- Wasternack C, Song S. Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J Exp Bot. 2017;68:1303–21.
- Browse J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol. 2009;60:183–205. doi:10.1146/annurev.arplant.043008.092007.
- Huang H, Liu B, Liu L, Song S. Jasmonate action in plant growth and development. J Exp Bot. 2017;68:1349–59. doi:10.1093/jxb/erw495.
- Goossens J, Fernandez-Calvo P, Schweizer F, Goossens A. Jasmonates: signal transduction components and their roles in environmental stress responses. Plant Mol Biol. 2016;91:673–89. doi:10.1007/s11103-016-0480-9.
- Wasternack C, Forner S, Strnad M, Hause B. Jasmonates in flower and seed development. Biochimie. 2013;95:79–85. doi:10.1016/j.biochi.2012.06.005.
- Zhou M, Memelink J. Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol Adv. 2016;34:441–9. doi:10.1016/j.biotechadv.2016.02.004.
- Shan X, Zhang Y, Peng W, Wang Z, Xie D. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J Exp Bot. 2009;60:3849–60. doi:10.1093/jxb/erp223.
- Yan C, Xie D. Jasmonate in plant defence: sentinel or double agent? Plant Biotechnol J. 2015;13:1233–40. doi:10.1111/pbi.12417.
- Zhang L, Zhang F, Melotto M, Yao J, He SY. Jasmonate signaling and manipulation by pathogens and insects. J Exp Bot. 2017;68:1371–85.
- Campos ML, Kang JH, Howe GA. Jasmonate-triggered plant immunity. J Chem Ecol. 2014;40:657–75. doi:10.1007/s10886-014-0468-3.
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–4. doi:10.1126/science.280.5366.1091.
- Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, Cheng Z, Peng W, Luo H, Nan F, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. The Plant Cell. 2009;21:2220–36. doi:10.1105/tpc.109.065730.
- Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–71. doi:10.1038/nature06006.
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–5. doi:10.1038/nature05960.
- Yan YX, Stolz S, Chetelat A, Reymond P, Pagni M, Dubugnon L, Farmer EE. A downstream mediator in the growth repression limb of the jasmonate pathway. The Plant Cell. 2007;19:2470–83. doi:10.1105/tpc.107.050708.
- Goossens J, Mertens J, Goossens A. Role and functioning of bHLH transcription factors in jasmonate signalling. J Exp Bot. 2017;68:1333–47.
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004;40:22–34. doi:10.1111/j.1365-313X.2004.02183.x.
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–27. doi:10.1111/j.1365-313X.2007.03373.x.
- Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70. doi:10.1016/j.tplants.2004.12.011.
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D. The jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. The Plant Cell. 2011;23:1795–814. doi:10.1105/tpc.111.083261.
- Fernandez-Calvo P, Chini A, Fernandez-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. The Plant Cell. 2011;23:701–15. doi:10.1105/tpc.110.080788.
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20:735–47. doi:10.1093/molbev/msg088.
- Weinthal D, Tzfira T. Imaging protein-protein interactions in plant cells by bimolecular fluorescence complementation assay. Trends Plant Sci. 2009;14:59–63. doi:10.1016/j.tplants.2008.11.002.
- Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics. 2000;156:1349–62.
