ABSTRACT
The Elongator complex interacts with RNA polymerase II and via histone acetylation and DNA demethylation facilitates epigenetically the transcription of genes involved in diverse processes in plants, including growth, development, and immune response. Recently, we have shown that the Elongator complex promotes hypocotyl elongation and photomorphogenesis in Arabidopsis thaliana by regulating the photomorphogenesis and growth-related gene network that converges on genes implicated in cell wall biogenesis and hormone signaling. Here, we report that germination in the elo mutant was delayed by 6 h in the dark when compared to the wild type in a time lapse and germination assay. A number of germination-correlated genes were down-regulated in the elo transcriptome, suggesting a transcriptional regulation by Elongator. We also show that the hypocotyl elongation defect observed in the elo mutants in darkness originates very early in the post-germination development and is independent from the germination delay.
The Elongator complex promotes the RNA polymerase II-mediated transcript elongation through epigenetic histone acetyl transferase (HAT) and DNA demethylation activities conferred by one of its six subunits, designated ELP3.Citation1-5 In plants, the complex regulates growth, development, immune response, sensitivity to drought and abscisic acid.Citation6 Elongator and genes are expressed in meristematic tissues, correlating with delayed growth, shortened primary roots, reduced lateral root density, abnormal leaves, defective inflorescence phyllotaxis, and reduced apical dominance in the elo mutants of Arabidopsis thaliana.Citation2,Citation7,Citation8 Germination delay of the elo3 mutant had already been mentionedCitation1 as well as severe perturbation in post-germinative growth that could be rescued by addition of sucrose in the growth media.Citation7 Nevertheless, a possible correlation between germination defects and defective seedling growth had not been investigated. Phenotypes of the elo mutants are frequently associated with reduced expression and histone acetylation of the genes implicated in processes affected by the elo mutations.Citation2,Citation4,Citation8-11 Recently, we have proposed a model in which Elongator represses the plant immune response and promotes hypocotyl elongation and photomorphogenesis via the transcriptional control of positive photomorphogenesis regulators and a growth-regulatory network that converges on genes involved in cell wall biogenesis and hormone signaling.Citation12 Here, we show that the elo3-6 mutants are delayed in germination independently from the lag in the very early post-germination growth, resulting in the hypocotyl elongation phenotype in the dark. We also identify germination-related genes down-regulated in the transcriptome of the mutant relative to the wild type.
To analyze the role of Elongator in the early development of Arabidopsis, we compared the germination of the elo3-6 (GABI-KAT collection code GABI555_H06)Citation2 mutant and the wild type (Columbia-0 [Col-0] accession) in the presence or absence of sucrose by means of a time lapse experiment. The experiments were repeated twice with independent seed batches of Col-0 and elo3-6. Per experiment, seed batches were harvested from plants grown at the same time under the same conditions. Seeds sterilized for 10 min in 5% (v/v) bleach with 0.05% (v/v) Tween 20 were washed in water, sown on half-strength MS mediumCitation13 with or without 1% (w/v) sucrose, and stratified at 4°C for 48 h. Seeds were illuminated for 6 h in white light (100 µmol m−2 s−1) to induce germination, transferred to darkness for the indicated time at 21°C to follow germination in a time lapse experiment with pictures taken every 1 hCitation14,Citation15 and with an infrared light of 930 nm for the imaging. Root radicle emergence of at least 10 out of 20 seeds was used as a proxy for germination and was delayed in the elo3-6 mutant by 6 h () in the presence or absence of sucrose. To identify the exact germination and early post-germination stage(s) delayed in the elo3-6 mutant,Citation16 stratified seeds were illuminated with white light and placed in the dark for germination. Samples of 50 Col-0 and elo3-6 seeds were taken every 3 h starting at 16 h in darkness. Lack of germination, testa rupture, endosperm rupture, and post-germination stages A to F were assessed under the stereomicroscope until 40 h. The number of seeds representing each developmental stage at a given time point was plotted (). Germination, defined as the point when more than 50% of the seeds reached endosperm rupture, was delayed by 6 h in elo3-6 (time point 25 h) relative to Col-0 (time point 19 h). The post-germination stages A to F were also achieved by elo3-6 plants with a delay of approximately 6 h, the stage assumed when at least 50% of the seedlings had reached this or a more advanced stage. At 31 h, almost all seeds of both genotypes had germinated (). After 40 h, more than 60% of the Col-0 seedlings had reached stage F, while the majority of the elo3-6 mutants remained at stages B to E and only approximately 30% were at stage F.
Figure 1. Germination and post-germination development of the elo3-6 mutant. (A) The Col-0 and elo3-6 seeds were sown on half-strength MS with 1% (w/v) sucrose, stratified for 48 h at 4°C, exposed to white light for 6 h to stimulate germination, shifted to darkness, and followed in time lapse with pictures taken every 1 h. The indicated time points refer to time after white light exposure. (B) Germination and post-germination stage percentage of Col-0 and elo3-6 seeds in darkness. The seeds were sown on half-strength MS without sucrose, stratified for 48 h at 4°C, exposed to white light for 6 h, and shifted to darkness. Fifty seeds were taken at the indicated time points. No germination, germination, and post-germination stages were observed under the stereomicroscope and quantified. (C) Col-0 and elo3-6 seedlings were grown 4 days on half-strength MS medium with (+) or without (−) sucrose in darkness. Bars represent mean values ± s.d. Differences between mutant and wild type were statistically analyzed with an unpaired two-tailed Student's t-test and significant differences are indicated with asterisks (P<0.05).
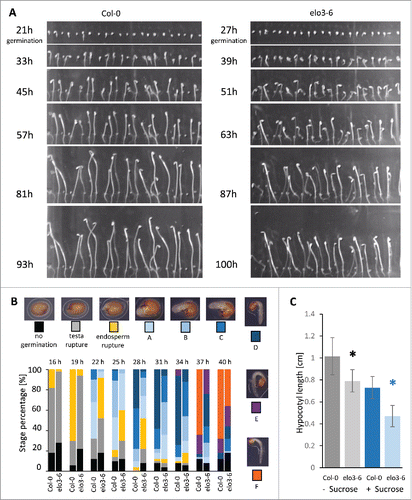
By means of the time lapse imaging, we followed the post-germination growth of the Col-0 and elo3-6 seedlings in darkness for 100 h. In addition to the germination delay, also the hypocotyl elongation was found to be postponed in the mutant seedlings. The hypocotyls of the elo3-6 plants were visibly shorter than those of the wild type already 24 h after germination (time points 45 h and 51 h for Col-0 and elo3-6, respectively) (). The delay in hypocotyl elongation in the dark for the elo3-6 seedlings grown for 3–7 days without sucrose has already been described.Citation12 Here, we show that this effect originates in early post-germinative growth and is independent from the germination defect. As the germination delay occurred both in the presence and absence of 1% (w/v) sucrose, we checked how sucrose affected the elongation of the elo3-6 hypocotyls. We compared the hypocotyl length of Col-0 and elo3-6 seedlings grown in the dark on media with and without sucrose. Under both conditions, the hypocotyls of elo3-6 were shorter than those of Col-0, but sucrose had an inhibitory effect on the hypocotyl elongation of both genotypes ().
As the Elongator complex regulates transcription epigenetically, we searched the microarray dataset of 4-day-old darkness-grown elo3-6 and Col-0 seedlings.Citation12 We found that the germination-related genes,Citation17 DELAY OF GERMINATION 1 (DOG1), HOMEOBOX-LEUCINE ZIPPER PROTEIN B-15 (ATHB15), VASCULAR-RELATED NAC-DOMAIN 2 (VND2), Oct-BINDING FACTOR (OBF)-BINDING PROTEIN 1 (OBP1), TEOSINTE BRANCHED1/CYCLOIDEA/PCF 14 (TCP14) and TPC15, DNA BINDING WITH ONE FINGER 5.6 (DOF5.6), DOF6, and DOF AFFECTING GERMINATION 2 (DAG2), were down-regulated in elo3-6 (). The gene coding for the positive regulator of light-induced germination, LONG HYPOCOTYL IN FAR-RED 1 (HFR1),Citation18,Citation19 was also down-regulated in elo3-6 and targeted by Elongator for histone acetylation.Citation12
Table 1. Germination-related genes down-regulated in 4-day-old seedlings of elo3-6. Gene were down-regulated at log2FC ≤ −0.5, corresponding to FC ≤ 0.7, P < 0.05. Data are available at NCBI, Gene Expression Omnibus, accession number GSE42053.
Although the Elongator complex expressed in meristems is a well-known regulator of many aspects of growth and development,Citation2,Citation6 its role in germination had only been suggested.Citation1 Our detailed analysis indicates that germination is delayed in the elo3-6 mutant, possibly as a result of transcriptional down-regulation of known germination regulators, and that this defect does not depend on the delay in early post-germination growth. Consequently, we propose a novel function of the Elongator complex in the control of the germination.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Martine De Cock for help in preparing the manuscript.
Additional information
Funding
References
- Nelissen H, Fleury D, Bruno L, Robles P, De Veylder L, Traas J, Micol JL, Van Montagu M, Inzé D, Van Lijsebettens M. The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc Natl Acad Sci U S A. 2005;102:7754–7759. doi:10.1073/pnas.0502600102.
- Nelissen H, De Groeve S, Fleury D, Neyt P, Bruno L, Bitonti B, Vandenbussche F, Van Der Straeten D, Yamaguchi T, Tsukaya H, et al. Plant Elongator regulates auxin-related genes during RNA polymerase II transcription elongation. Proc Natl Acad Sci U S A. 2010;107:1678–1683. doi:10.1073/pnas.0913559107.
- Glatt S, Müller CW. Structural insights into Elongator function. Curr Opin Struct Biol. 2013;23:235–242. doi:10.1016/j.sbi.2013.02.009.
- DeFraia CT, Wang Y, Yao J, Mou Z. Elongator subunit 3 positively regulates plant immunity through its histone acetyltransferase and radical S-adenosylmethionine domains. BMC Plant Biol. 2013;13:102. doi:10.1186/1471-2229-13-102.
- Woloszynska M, Le Gall S, Van Lijsebettens M. Plant Elongator-mediated transcriptional control in a chromatin and epigenetic context. Biochim Biophys Acta. 2016;8:1025–1033. doi:10.1016/j.bbagrm.2016.06.008.
- Ding Y, Mou Z. Elongator and its epigenetic role in plant development and responses to abiotic and biotic stresses. Front Plant Sci. 2016;6:296. doi:10.3389/fpls.2015.00296.
- Skylar A, Matsuwaka S, Wu X. ELONGATA3 is required for shoot meristem cell cycle progression in Arabidopsis thaliana seedlings. Dev Biol. 2013;382:436–445.doi:10.1016/j.ydbio.2013.08.008.
- Jia Y, Tian H, Li H, Yu Q, Wang L, Friml J, Ding Z. The Arabidopsis thaliana elongator complex subunit 2 epigenetically affects root development. J Exp Bot. 2015;66:4631–4642. doi:10.1093/jxb/erv230.
- An C, Wang C, Mou Z. The Arabidopsis Elongator complex is required for nonhost resistance against the bacterial pathogens Xanthomonas citri subsp. citri and Pseudomonas syringae pv. phaseolicola NPS3121. New Phytol. 2017;214:1245–1259. doi:10.1111/nph.14442.
- Wang C, Ding Y, Yao J, Zhang Y, Sun Y, Colee J, Mou Z. Arabidopsis Elongator subunit 2 positively contributes to resistance to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola. Plant J. 2015;83:1019–1033. doi:10.1111/tpj.12946.
- Wang Y, An C, Zhang X, Yao J, Zhang Y, Sun Y, Yu F, Amador DM, Mou Z. The Arabidopsis Elongator complex subunit2 epigenetically regulates plant immune responses. Plant Cell. 2013;25:762–776. doi:10.1105/tpc.113.109116.
- Woloszynska M, Gagliardi O, Vandenbussche F, De Groeve S, Alonso Baez L, Neyt P, Le Gall S, Fung J, Mas P, Van Der Straeten D, Van Lijsebettens M. The Elongator complex regulates hypocotyl growth in darkness and during photomorphogenesis. J Cell Sci. 2018. doi:10.1242/jcs.203927.
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi:10.1111/j.1399-3054.1962.tb08052.x.
- Vandenbussche F, Petrášek J, Žádníková P, Hoyerová K, Pešek B, Raz V, Swarup R, Bennett M, Zažimalová E, Benková E, et al. The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development. 2010;137:597–606. doi:10.1242/dev.040790.
- Smet D, Žádníková P, Vandenbussche F, Benková E, Van Der Straeten D. Dynamic infrared imaging analysis of apical hook development in Arabidopsis: the case of brassinosteroids. New Phytol. 2014;202:1398–1411. doi:10.1111/nph.12751.
- Müller K, Tintelnot S, Leubner-Metzger G. Endosperm-limited Brassicaceae seed germination: Abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol. 2006;47:864–877. doi:10.1093/pcp/pcj059.
- Narsai R, Gouil Q, Secco D, Srivastava A, Karpievitch YV, Liew LC, Lister R, Lewsey MG, Whelan J. Extensive transcriptomic and epigenomic remodelling occurs during Arabidopsis thaliana germination. Genome Biol. 2017;18:172, doi:10.1186/s13059-017-1302-3.
- Shi H, Zhong S, Mo X, Liu N, Nezames CD, Deng XW. HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell. 2013;25, 3770–3784. doi:10.1105/tpc.113.117424.
- Shi H, Wang X, Mo X, Tang C, Zhong S, Deng XW. Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. Proc Natl Acad Sci U S A. 2015;112:3817–3822. doi:10.1073/pnsa.1502405112.
