ABSTRACT
Autophosphorylation of PRR is a critical event for the activation of immune signaling in plant. However, the detailed function of these phosphorylation sites is still not well understood. We analyzed the function of an autophosphorylation site of Arabidopsis CERK1, Y428, in immune signaling. Biochemical characterization of CERK1 mutants transiently expressed in N. benthamiana indicated that Y428 plays a crucial role for the in vivo activation of CERK1, differently from the previous observation by the in vitro kinase assay with its cytoplasmic domain. Similar discrepancy between in vitro and in vivo kinase assay was also reported for the corresponding phosphorylation site of EFR, suggesting that these conserved tyrosine residues play important roles for the activation of both RD and non-RD RLKs.
Plants have the ability to detect invading microbes through the recognition of microbe-associated molecular patterns (MAMPs) and initiate various immune responses.Citation1,Citation2 Cell surface pattern-recognition receptors (PRRs), which consist of receptor-like proteins and receptor-like kinases (RLKs), recognize the MAMPs and activate downstream immune signaling.Citation3 Although the MAMP-triggered autophosphorylation of RLKs is known to be critical for the activation of downstream signaling,Citation4 detailed function of each autophosphorylation site in a specific RLK is still not well understood.
The Arabidopsis LysM-RLK CERK1 mediates immune responses triggered by chitin and peptidoglycan.Citation5-7 It has been shown that the kinase activity of CERK1 is essential for the activation of immune responses and CERK1 itself is phosphorylated by chitin treatment.Citation8,Citation9 Petutschnig et al. identified four or five in vivo phosphorylation sites of CERK1 and showed that the phosphorylation of at least three of them was dependent on chitin treatment.Citation8 Recently, we also identified 41 in vitro and 15 in vivo phosphorylation sites of CERK1 and indicated that at least three of them play important roles in chitin signaling based on the complementation experiments with the CERK1 mutants of these phosphorylation sites.Citation9 Among them, T479A and Y428F mutants completely lost the ability to complement the cerk1 mutant for chitin responses. Mutation of T573A also showed a much less complementation ability compared to the wild type CERK1. In vitro kinase assay with the kinase domains expressed in E. coli showed that the T479A mutant almost completely lost autophosphorylation activity and the T573A mutant also showed a reduced autophosphorylation activity, indicating T479 and T573 contribute to chitin signaling through the regulation of the kinase activity of CERK1. On the other hand, the mutation of Y428 did not affect the in vitro kinase activity, indicating Y428 phosphorylation might contribute to chitin signaling through a different mechanism.
To obtain further insight into the function of Y428 phosphorylation, we evaluated the effect of this mutation on the biological activity and the in vivo phosphorylation of CERK1 by using transient overexpression of the Y428F mutant in Nicotiana benthamiana. Previously, Pietraszewska-bogiel et al. reported that the overexpression of CERK1 in N. benthamiana induced defense-like responses including cell death.Citation10 They showed that these responses were dependent on CERK1 kinase activity.Citation10 We also reported that the CERK1 expressed in N. benthamiana was autophosphorylated in the absence of chitin treatment.Citation11 These results indicated that the transient expression in N. benthamiana is useful for the evaluation of biological activity as well as in vivo phosphorylation status of CERK1.
Transient overexpression of wild type CERK1 in N. benthamiana leaves induced cell death () as previously reported.Citation10 On the other hand, the expression of Y428F mutant of CERK1 did not induce cell death as similar to the kinase-dead CERK1(D441V). These results again confirmed that the autophosphorylation site Y428 is essential for the function of CERK1 leading to the defense-like responses such as cell death.
Figure 1. Evaluation of cell death induced by the overexpression of CERK1 constructs in N. Benthamiana. N. benthamiana was grown under a photoperiod of 14 h of light (20°C) and 10 h of darkness (22°C). A. tumefaciens C58C1 carrying the corresponding construct was infiltrated into N. benthamiana leaves using a syringe. Cell death induced by the overexpression of CERK1 constructs was monitored at 4 days after infiltration. (A) Macroscopic observation, (B) observation after decoloration with ethanol : acetic acid (3 : 1). The plasmids for transient expression in N. benthamiana were constructed as follows and finally inserted into pGWB14.Citation17 pENTR/D-TOPO CERK1(D441V) and CERK1(Y428F) were generated as reported previously.Citation9 pENTR/D-TOPO CERK1(Y428D) and CERK1(Y428E) were amplified from pENTR/D-TOPO CERK1 using PrimeSTAR Max DNA Polymerase with primer pairs 5′-TTTAGAAGATATCCACGAGCACACGG-3′/5′-TGGATATCTTCTAAACCTCTAGCTGAGTCTAGT-3′ and 5′-TTTAGAAGAAATCCACGAGCACACGG-3′/5′-GTGGATTTCTTCTAAACCTCTAGCTGAGTCTAG-3′.
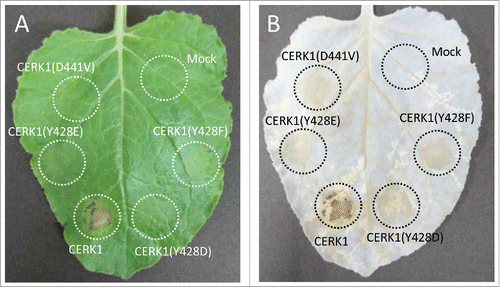
The fact that the Y428F mutant of CERK1 failed to induce cell death in N. benthamiana as similar to the kinase-dead CERK1 raised a question on its in vivo phosphorylation status. Thus, the phosphorylation status of the HA-tagged CERK1 expressed in N. benthamiana was analyzed by western blotting combined with SDS-PAGE (). The band of the wild type CERK1-3HA showed a clear upward shift compared to the kinase-dead mutant. This band shift was previously confirmed to show the autophosphorylation of CERK1.Citation9 On the other hand, the band of CERK1(Y428F)-3HA did not show such an upward shift, indicating that the mutant CERK1 was not phosphorylated in N. benthamiana. These observations are different from the results of previous in vitro kinase assay where the heterologously expressed kinase domain of this mutant was normally autophosphorylated and indicated that Y428 plays a pivotal role in the in vivo activation of CERK1 kinase required for downstream defense signaling. These results also suggest that the experiments with the soluble kinase domains do not always predict the behavior of the full-length, membrane bound RLKs. One possible scenario that explains such a discrepancy between in vitro and in vivo experiments could be the heterologously expressed kinase domain of CERK1(Y428F) is freely diffusible in the in vitro experiment and phosphorylates with each other through an intermolecular mechanismCitation9 but the activation of full length CERK1 fixed to the plasma membrane takes a more complicated process that requires the phosphorylation or the presence of Y428.
Figure 2. Evaluation of the phosphorylation status of CERK1 expressed in N. benthamiana. Tobacco leaves expressing CERK1 were collected 24h after infiltration. Microsomal fraction was prepared from the leaves and solubilized with TBS containing 1% TritonX-100 as reported previously.Citation18 Samples corresponding to 10 µg protein were subjected to SDS-PAGE and detected by western blotting with anti-HA antibody (SIGMA).
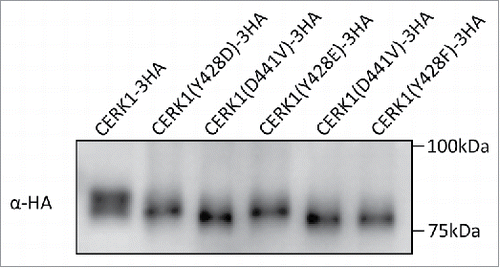
To further evaluate the contribution of the phosphorylation of Y428 for the regulation of CERK1 kinase activity in vivo, we constructed phosphomimetic mutants of CERK1 by substituting Y428 with Asp or Glu and evaluated their function in N. benthamiana. Both of the phosphomimetic mutants did not induce cell death in the N. benthamiana leaves as similar to the kinase-dead construct (). On the other hand, the bands of CERK1(Y428D)-3HA and CERK1(Y428E)-3HA expressed in N. benthamiana showed a slight upward shift in the SDS-PAGE (). These results suggested that the phosphomimetic mutants of CERK1 autophosphorylated themselves only partially in tobacco and could not induce immune responses, such as cell death.
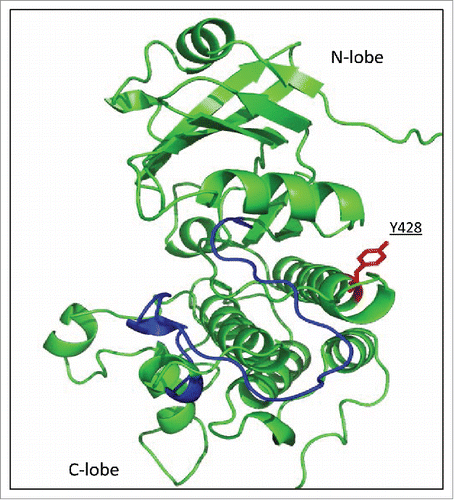
As the phosphomimetic mutants failed to recover the function of wild type CERK1 in tobacco, except a slight autophosphorylation, it is difficult to conclude whether the phosphorylation of Y428 is important for the in vivo activation of CERK1. On the other hand, it seems evident that the substitution with these acidic residues does not always mimic the function of real phosphorylation sites, especially in the case of phosphotyrosine residue that differs from Asp or Glu for the presence of bulky, hydrophobic aromatic ring. Limitation of the use of phosphomimetic approach has also been described so far.Citation12,Citation13 Y428 of CERK1 is located on the α-helix in the C-terminal lobe () and the phosphorylation of this residue might affect the overall structure of CERK1 through the lobe interactions, though it remains to be clarified based on the detailed structural studies of the kinase domain.
Figure 3. Location of Y428 on the molecular model of CERK1 kinase domain. Homology model of the CERK1 kinase domain was built as reported previously.Citation9 Activation loop was shown in blue and Y428 was shown in red.
Tyrosine residues corresponding to Y428 of CERK1 are conserved in various RLKs including both LysM-RLKs and LRR-RLKs. Among them, Y836 of EFR, the receptor of bacterial elongation factor Tu (EF-Tu), is phosphorylated upon perception of elf18 and essential for elf18-induced ROS generation.Citation14 It was reported that the mutation of Y836F did not affect the in vitro autophosphorylation of the EFR kinase domain, as similar to CERK1(Y428F) mutant. Based on the observation, it was concluded that the Y836 residue is not required for the overall kinase activity of EFR.Citation15 However, careful observation of the result of the in vivo phosphorylation analysis reported in this paper (Macho et al., Fig. S914) indicated, though not discussed in the paper, that the elf18-induced phosphorylation of Y836F mutant was decreased compared to the wild type EFR, indicating that Y836 is also important for the in vivo activation of EFR. It is interesting that the conserved tyrosine residues in both RD (CERK1) and non-RD (EFR) kinases, which are distinguished based on the presence (RD) or absence (non-RD) of an arginine residue preceding the catalytic aspartic residue and also different in the kinase activity as well as the activation mechanism,Citation16 seem to play an important role in the regulation of in vivo kinase activity, though the detailed molecular mechanism remains to be clarified.
Abbreviations
| CERK1 | = | chitin elicitor receptor kinase |
| MAMP | = | microbe-associated molecular pattern |
| LysM | = | lysin motif |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Yoshitake Desaki for helpful advices. We indebted to Dr. Zui Fujimoto of NARO for the generation of the molecular model of CERK1 kinase domain. We also thank Ms. Noriko Motoyama, Ms. Hikaru Shimada and Mr. Masatoshi Shibuya for making CERK1(D441V) and CERK1(Y428F) entry clones.
Additional information
Funding
References
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi:10.1146/annurev.arplant.57.032905.105346.
- Couto D, Zipfel C. Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol. 2016;16:537–52. doi:10.1038/nri.2016.77.
- Tang D, Wang G, Zhou JM. Receptor Kinases in Plant-Pathogen interactions: More than pattern recognition. Plant Cell. 2017;29:618–37. doi:10.1105/tpc.16.00891.
- Macho AP, Zipfel C. Plant PRRs and the activation of innate immune signaling. Mol Cell. 2014;54:263–72. doi:10.1016/j.molcel.2014.03.028.
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104:19613–8. doi:10.1073/pnas.0705147104.
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20:471–81. doi:10.1105/tpc.107.056754.
- Willmann R, Lajunen HM, Erbs G, Newman MA, Kolb D, Tsuda K, Katagiri F, Fliegmann J, Bono JJ, Cullimore JV. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc Natl Acad Sci U S A. 2011;108:19824–9. doi:10.1073/pnas.1112862108.
- Petutschnig EK, Jones AM, Serazetdinova L, Lipka U, Lipka V. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J Biol Chem. 2010;285:28902–11. doi:10.1074/jbc.M110.116657.
- Suzuki M, Shibuya M, Shimada H, Motoyama N, Nakashima M, Takahashi S, Suto K, Yoshida I, Matsui S, Tsujimoto N. Autophosphorylation of specific Threonine and Tyrosine residues in Arabidopsis CERK1 is essential for the activation of Chitin-Induced Immune signaling. Plant Cell Physiol. 2016;57:2312–22. doi:10.1093/pcp/pcw150.
- Pietraszewska-Bogiel A, Lefebvre B, Koini MA, Klaus-Heisen D, Takken FL, Geurts R, Cullimore JV, Gadella TW. Interaction of Medicago truncatula lysin motif receptor-like kinases, NFP and LYK3, produced in Nicotiana benthamiana induces defence-like responses. PLoS One. 2013;8:e65055. doi:10.1371/journal.pone.0065055.
- Shinya T, Yamaguchi K, Desaki Y, Yamada K, Narisawa T, Kobayashi Y, Maeda K, Suzuki M, Tanimoto T, Takeda J. Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J. 2014;79:56–66. doi:10.1111/tpj.12535.
- Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–35. doi:10.1016/j.devcel.2008.06.011.
- Chen X, Chern M, Canlas PE, Jiang C, Ruan D, Cao P, Ronald PC. A conserved threonine residue in the juxtamembrane domain of the XA21 pattern recognition receptor is critical for kinase autophosphorylation and XA21-mediated immunity. J Biol Chem. 2010;285:10454–63. doi:10.1074/jbc.M109.093427.
- Macho AP, Schwessinger B, Ntoukakis V, Brutus A, Segonzac C, Roy S, Kadota Y, Oh MH, Sklenar J, Derbyshire P. A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science. 2014;343:1509–12. doi:10.1126/science.1248849.
- Macho AP, Lozano-Duran R, Zipfel C. Importance of tyrosine phosphorylation in receptor kinase complexes. Trends Plant Sci. 2015;20:269–72. doi:10.1016/j.tplants.2015.02.005.
- Dardick C, Ronald P. Plant and animal pathogen recognition receptors signal through non-RD kinases. PLoS Pathog. 2006;2:e2. doi:10.1371/journal.ppat.0020002.
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104:34–41. doi:10.1263/jbb.104.34.
- Shinya T, Motoyama N, Ikeda A, Wada M, Kamiya K, Hayafune M, Kaku H, Shibuya N. Functional characterization of CEBiP and CERK1 homologs in arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol. 2012;53:1696–706. doi:10.1093/pcp/pcs113.
