ABSTRACT
Abscisic acid (ABA) plays critical roles during plant growth and development in response to various stresses. Arabidopsis thaliana histone demethylases JUMONJI-C DOMAIN-CONTAINING PROTEIN 30 (JMJ30) and JMJ32 control ABA-mediated growth arrest during the post-germination stage (2–3 days after germination). However, the roles of JMJ30 and JMJ32 in ABA responses at later stages of plant development remain largely unknown. Here, we show that JMJ30 and JMJ32 mediate ABA responses during root development. In the presence of ABA, jmj30 jmj32 double mutants display longer primary roots than the wild type. Loss-of-function mutation in the SNF1-RELATED PROTEIN KINASE 2.8 (SnRK2.8) gene also led to a longer primary root phenotype in response to ABA. Analysis of JMJ30/JMJ32 and SnRK2.8 expression suggested that they act in the same pathway to mediate ABA responses during root elongation at the seedling stage. Our findings highlight the importance of the JMJ30/JMJ32-SnRK2.8 module at two different developmental stages.
External stress negatively impacts growth, development, and productivity of plants.Citation1 One major stress is categorized as abiotic or environmental stress, such as unfavorable atmosphere, chemical elements, sunlight/temperature, wind and water. Because plants are sessile organisms, they have developed various mechanisms to protect themselves against stresses. In the last two decades, much research has focused on understanding plant molecular frameworks toward improving crop yield even under stress conditions.Citation2-Citation4
Abscisic acid (ABA) is a key stress-signaling hormone.Citation5,Citation6 In response to stress such as water deficit and high salt, not only the amount of ABA, but also ABA perception and response are modulated. Osmotic stress caused by drought and high salt triggers ABA biosynthesis, and the resulting ABA accumulates in the cytosol and binds to the ABA receptors PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE(PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS (RCAR).Citation7,Citation8 The activated ABA receptors bind to type 2C protein phosphatases (PP2Cs) like ABSCISIC ACID-INSENSITIVE1 (ABI1) or ABI2, inhibiting the catalytic activity of PP2C.Citation9 SNF1-RELATED PROTEIN KINASE2 (SnRK2) kinases are then released from PP2C-mediated inactivation and trigger gene expression through phosphorylation.Citation10-Citation12 After reaching a certain threshold of ABA concentration or signaling, stomata are closed and gene expression is changed through cis-acting ABA-responsive elements (ABREs).Citation13 ABRE and a group of ABRE-binding transcription factors have pivotal roles in ABA-dependent gene expression. Although ABA-dependent gene induction is well characterized, how it is controlled at the levels of histone modification remains unclear.
Histones function both positively and negatively in the regulation of gene expression.Citation14 The N-terminal tail of histone H3 is modified post-translationally through acetylation, phosphorylation, methylation and ubiquitination.Citation15 Histone modification enzyme complexes catalize reversible lysine methylation central to epigenetic regulation by specifying when, where and which histone residues need to be modified. Despite their importance, the role of histone modification enzymes in ABA responses is not well characterized.
We recently reported that the histone demethylases JUMONJI-C DOMAIN-CONTAINING PROTEIN 30 (JMJ30) and JMJ32 control ABA-mediated growth arrest during the post-germination stage.Citation16 Under unfavorable environmental conditions, the B3 domain transcription factor ABSCISIC ACID INSENSITIVE3 (ABI3) is activated by ABA.Citation17 ABA-activated ABI3 promotes expression of JMJ30, presumably by direct binding via the evolutionally conserved RY motif.Citation16 JMJ30 and JMJ32 then remove repressive H3K27me3 marks at the SnRK2.8 locus to activate its expression.Citation16 The upregulated SnRK2.8 promotes ABA-dependent gene expression, which feeds forward to ABI3 activation.Citation16
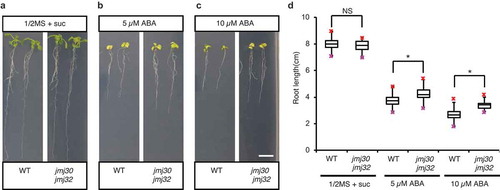
A comprehensive expression study of JMJ genes in response to stress revealed that JMJ30 is upregulated by ABA during the vegetative stage in Arabidopsis thaliana.Citation18 However, the function of JMJ30 and JMJ32 in the ABA response during the vegetative stage remains unknown. To understand their roles, we performed phenotypic analysis using jmj30 jmj32 double mutants in the absence and presence of ABA at the vegetative stage (–). Three-day-old wild-type and jmj30-2 jmj32-1 double mutant seedlings were transferred to half-strength MS plates with or without ABA. When grown and maintained on half-strength MS plates without ABA, wild-type and jmj30-2 jmj32-1 plants showed no obvious phenotypic differences (); both displayed leaves of normal size and color and well-grown primary roots with many lateral roots (). No significant difference in primary root length was observed between the wild type and jmj30-2 jmj32-1 without ABA (p > .05 by two-tailed Student’s t-test) (). ABA-treated plants of both genotypes had smaller and paler leaves and shorter roots compared with control plants (–). In the presence of 5 µM ABA, primary root length in the wild type was 3.7 ± 0.1 cm while roots of jmj30-2 jmj32-1 plants were significantly longer at 4.2 ± 0.1 cm (p < .01 by two-tailed Student’s t-test). Root elongation was inhibited more in the presence of 10 µM ABA than 5 µM ABA (); however, there were still significant differences in root length between the wild type and jmj30-2 jmj32-1 (p < .01 by two-tailed Student’s t-test) (). These results suggest that JMJ30 and JMJ32 are required for ABA-dependent inhibition of root growth during the vegetative stage.
Figure 1. Root elongation in jmj30 jmj32 double mutants is less sensitive to ABA. (A–C) Representative images of wild-type (WT) and jmj30-2 jmj32-1 plants in the absence and presence of ABA. Wild-type and jmj30-2 jmj32-1 seeds were sown on half-strength MS with 1% sucrose and stratified at 4°C for 3 days. Plants were grown under 24 h of light for 3 days and then transplanted onto half-strength MS plates with 1% sucrose supplemented with 0 µM ABA (A), 5 µM ABA (B), or 10 µM ABA (C) and grown vertically under 24 h of light for an additional 7 days. Bar = 1 cm. (D) Quantification of root length in wild-type and jmj30-2 jmj32-1 plants shown in (A–C). Asterisks indicate significant differences based on two-tailed Student’s t-test; p < .01; NS, nonsignificant. Values represent mean ± SD of 24 plants.
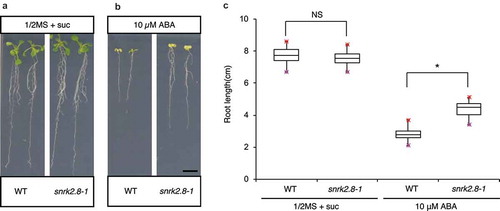
To understand the role of SnRK2.8 in ABA-mediated root elongation at the vegetative stage, we conducted phenotypic analyses of wild-type and snrk2.8–1 plants. There was no significant difference in phenotype between wild-type and snrk2.8–1 plants when grown and maintained on half-strength MS plates without ABA (p > .05 by two-tailed Student’s t-test) (). When transferred onto 10 µM ABA plates, root growth was inhibited in both the wild type and snrk2.8–1 (). However, the snrk2.8–1 mutant was less sensitive to ABA, similar to jmj30-2 jmj32-1 double mutants (p < .01 by two-tailed Student’s t-test) (). These results suggest that SnRK2.8 is required for ABA-dependent inhibition of root growth during the vegetative stage.
Figure 2. Root elongation in snrk2.8 mutants is less sensitive to ABA. (A, B) Representative images of wild-type (WT) and snrk2.8–1 plants in the absence and presence of ABA. Wild-type and snrk2.8–1 seeds were sown on half-strength MS with 1% sucrose and stratified at 4°C for 3 days. Plants were grown under 24 h of light for 3 days and then transplanted onto half-strength MS plates with 1% sucrose supplemented with 0 µM ABA (A) or 10 µM ABA (B) and grown vertically under 24 h of light for an additional 7 days. Bar = 1 cm. (C) Quantification of root length in wild-type and snrk2.8–1 plants shown in (A, B). Asterisk indicates significant difference based on two-tailed Student’s t-test; p< .01; NS, non-significant. Values represent mean ± SD of 24 plants.
To examine the relationship between JMJ30/JMJ32 and SnRK2.8 in response to ABA during the vegetative stage, we conducted reverse-transcription quantitative polymerase chain reaction (RT-qPCR) analysis (–). JMJ30 is upregulated during the postgermination stage by ABI3 in response to ABA.Citation16 To understand JMJ30 and JMJ32 expression in response to ABA during the vegetative stage, we first examined JMJ30 and JMJ32 expression levels (). Consistent with previous observations, JMJ30 was upregulated in response to ABA (p < .01 by two-tailed Student’s t-test) (). Similar to the postgermination stage, upregulation of JMJ32 expression was not observed (). To further confirm whether JMJ30 upregulation in response to ABA is dependent on ABI3 function, we examined ABI3 expression at the vegetative stage. Although we observed a significant difference in ABI5 expression in response to ABA, there was no difference in expression of the ABI3 gene (). These data suggest that JMJ30 is upregulated by a factor other than ABI3 in vegetative stage, unlike in post-germination stage.
Figure 3. JMJ30 and SnRK2.8 expression is induced by ABA. (A–D) Expression of JMJ30 (A), JMJ32 (B), ABI3 (C), and ABI5 (D) in wild-type (WT) plants in response to 10 µM ABA. Results are from three independent experiments. Values represent mean ± SEM. Asterisks indicate significant differences based on two-tailed Student’s t-test; p < .01; NS, nonsignificant. (E) Expression of SnRK2.8 in wild-type and jmj30-2 jmj32-1 plants in response to 10 µM ABA. Results are from three independent experiments. Values represent mean ± SEM. Asterisk indicates significant differences based on one-way ANOVA test; p < .01. Different letters indicate significant differences based on post-hoc Tukey’s HSD test; p < .01.
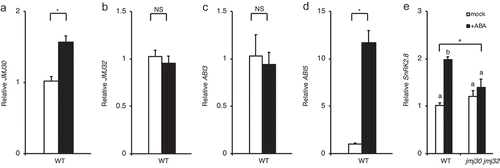
We next addressed the expression levels of SnRK2.8 (). ABA-treated wild-type plants had more SnRK2.8 transcripts than mock-treated wild-type plants (p < .01 by one-way ANOVA test) (WT with ABA vs. WT without ABA: p < .01 by post-hoc Tukey’s HSD) (). In addition, SnRK2.8 was not upregulated in the jmj30-2 jmj32-1 background with or without ABA treatment (jmj30-2 jmj32-1 with ABA vs. jmj30-2 jmj32-1 without ABA: p > .05 by post-hoc Tukey’s HSD) (). This result implies that SnRK2.8 expression is controlled by JMJ30 in response to ABA during the vegetative phase.
We previously showed that the function of the JMJ30/JMJ32-SnRK2.8 module is dependent on the ABA-dependent transcription factor ABI3 during the postgermination stage. Here, we demonstrated the role of the JMJ30/JMJ32-SnRK2.8 module in response to ABA during root elongation at the vegetative stage. Although the function of the JMJ30/JMJ32-SnRK2.8 module in response to ABA is well conserved between the two different developmental stages, the upstream regulators are different. Thus, we conclude that an unknown factor(s) – X(s) – activates JMJ30 in response to ABA during root elongation at the vegetative stage. It will be interesting to identify such a factor in the future.
Materials and methods
Plant materials and growth conditions
All Arabidopsis thaliana lines used in this study were in the Columbia (Col-0) background. The jmj30-2 jmj32-1 mutant was described previously.Citation19 The snrk2.8-1 (SALK_073395) mutant was obtained from the Arabidopsis Biological Resource Center (ABRC). Prior to growth, genotypes were confirmed by PCR using Emerald Amp polymerase (Takara). Primers for genotyping were as follows: jmj30-2 genotyping-FW, CAAACTCTGCTGCAATCGATTTC; jmj30-2 genotyping-RV, GAAAATGTCACAAGCTCTTGCTTC; jmj32-1 genotyping-FW, GACTGAGAAAACCTGAACTCAGC; jmj32-1 genotyping-RV, GTCGTGTAAAGGACTGAAGGTTG; snrk2.8-1 genotyping-FW, CAAACCATGACACATCAGCAC; snrk2.8-1 genotyping-RV, AGGCTCCTGTTAATCACCAGG. All plants were grown at 22°C in a growth chamber under continuous light conditions after stratification at 4°C for 3 days.
Phenotypic and statistical analyses
Procedures for preparation of half-strength MS plates and seed surface sterilization were described previously.Citation16 For root elongation assays, sterilized wild-type, jmj30-2 jmj32-1, and snrk2.8-1 seeds were placed on half-strength MS plates, stratified at 4°C for 3 days, and then placed in a growth chamber at 22°C under continuous light for 3 days. Three-day-old plants were transplanted onto half-strength MS plates with 1% sucrose supplemented with 0, 5, or 10 µM ABA and grown vertically under 24 h of light for an additional 7 days. Primary root length was measured, and statistical analyses were conducted using Microsoft Excel. Statistical significance was computed using a two-tailed Student’s t-test.
Expression analysis
For ABA treatment, 4-day-old stratified plants grown on half-strength MS plates with 1% sucrose were treated with 10 µM ABA to induce rapid changes in gene expression. After 3 h of treatment, seedlings were used for RNA extraction. RNA isolation and RT-qPCR methods followed a previously described protocol.Citation20 Three independent biological replicates were performed for qPCR analyses, and four technical replicates were conducted for each experiment. Statistical significance was computed using either one- way ANOVA test followed by post- hoc Tukey’s HSD test or two- tailed Student’s t-test for multiple- and single-paircomparisons, respectively. Primers for expression analyses were as follows: EIF4A1- FW, TCTTGGTGAAGCGTGATGAG; EIF4A1- AATCAACCTTACGCCTGGTG; JMJ30- FW GAATCACTTGGACTACCT CAATGC; JMJ30- RV, CATTGGAGACGATTTATT GGTCC; JMJ32- FW, GTTTCATTGTA CTGTCAAGGCTGG; JMJ32- RV, CATACTTGAT GTCAAACTGCA TGTC; ABI3- FW, ATGTATCTCCTCGAG AACAC; ABI3- RV, CCCTCGTATCAAATATTTG CC; ABI5- FW, ACCTAATCCAAACC CGAACC; ABI5- RV, TACCCTCCTCCTCCTGTCCT; SnRK2.8- FW, GTTGCCAACCCT GAAAAGAG; SnRK2.8- RV, CCGAGCTTCTTCAATGATCC.
Disclosure of potential conflicts of interest
No potential conflicts of interest are disclosed.
Additional information
Funding
References
- Pandey P, Irulappan V, Bagavathiannan MV, Senthil-Kumar M. Impact of combined abiotic and biotic stresses on plant growth and avenues for crop improvement by exploiting physio-morphological traits. Front Plant Sci. 2017;8:537. doi:10.3389/fpls.2017.00537.
- Mickelbart MV, Hasegawa PM, Bailey-Serres J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat Rev Genet. 2015;16:237. doi:10.1038/nrg3901.
- Fahad S, Bajwa AA, Nazir U, Anjum SA, Farooq A, Zohaib A, Sadia S, Nasim W, Adkins S, Saud S, et al. Crop production under drought and heat stress: plant responses and management options. Front Plant Sci. 2017;8:1147. doi:10.3389/fpls.2017.01147.
- Pereira A. Plant abiotic stress challenges from the changing environment. Front Plant Sci. 2016;7:1123. doi:10.3389/fpls.2016.01123.
- Verma V, Ravindran P, Kumar PP. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016;16:86. doi:10.1186/s12870-016-0796-2.
- Finkelstein R. Abscisic Acid synthesis and response. Arab B. 2013; 11:e0166–e0166. doi:10.1199/tab.0166.
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi:10.1146/annurev-arplant-042809-112122.
- Park S-Y, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068 LP–1071. Available from: http://science.sciencemag.org/content/324/5930/1068.abstract
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001;25:295–303. doi:10.1046/j.1365-313x.2001.00965.x.
- Kulik A, Wawer I, Krzywińska E, Bucholc M, Dobrowolska G. SnRK2 protein kinases–key regulators of plant response to abiotic stresses. OMICS. 2011;15:859–872. doi:10.1089/omi.2010.0113.
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, et al. Three arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50:1345–1363. doi:10.1093/pcp/pcp083.
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci. 2009;106:17588 LP–17593. doi:10.1073/pnas.0907095106.
- Mizoi J, Yoshida T, Fujita Y, Nakajima J, Ohori T, Todaka D, Nakashima K, Hirayama T, Shinozaki K, Yamaguchi-Shinozaki K. An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a trasnscription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 2011;52:2136–2146. doi:10.1093/pcp/pcp143.
- You Y, Sawikowska A, Neumann M, Posé D, Capovilla G, Langenecker T, Neher RA, Krajewski P, Schmid M. Temporal dynamics of gene expression and histone marks at the Arabidopsis shoot meristem during flowering. Nat Commun. 2017;8:15120. doi:10.1038/ncomms15120.
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi:10.1038/cr.2011.22.
- Wu J, Ichihashi Y, Suzuki T, Shibata A, Shirasu K, Yamaguchi N, Ito T. Abscisic acid-dependent histone demethylation during post-germination growth arrest in Arabidopsis. Plant Cell Environ. 2019. doi:10.1111/pce.13547.
- Kagaya Y, Okuda R, Ban A, Toyoshima R, Tsutsumida K, Usui H, Yamamoto A, Hattori T. Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in Arabidopsis. Plant Cell Physiol. 2005;46:300–311. doi:10.1093/pcp/pci031.
- Qian S, Wang Y, Ma H, Zhang L. Expansion and functional divergence of Jumonji C-containing histone demethylases: significance of duplications in ancestral angiosperms and vertebrates. Plant Physiol. 2015;168:1321 LP–1337. doi:10.1104/pp.15.00520.
- Gan E-S, Xu Y, Wong J-Y, Geraldine Goh J, Sun B, Wee W-Y, Huang J, Ito T. Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat Commun. 2014;5:5098. doi:10.1038/ncomms6098.
- Yamaguchi N, Huang J, Xu Y, Tanoi K, Ito T. Fine-tuning of auxin homeostasis governs the transition from floral stem cell maintenance to gynoecium formation. Nat Commun. 2017;8:1125. doi:10.1038/s41467-017-01252-6.
