ABSTRACT
Virus-induced silencing gene technology has been increasingly used; however, a controversy exists among researchers about whether using the sense or antisense orientation of the gene target is more efficient. Herein, instead of using the entire citrus tree, a reduced system consisting of a single leaf, 5 cm of the stem and a few roots was established to fairly compare between the sense or antisense orientation of phytoene desaturase gene (pds) in the Citrus tristeza virus vector, for improved RNAi efficiency. Although the virus titers were similar in the two cases, the gene expression of pds was significantly lower when using the antisense orientation than using the sense orientation. I hypothesize that the extra effect from use of antisense orientation is due to the role of subgenomic RNA as a supplemental source for complementary sequences, thus resulting in more RNAi.
Virus-induced gene silencing (VIGS) is a useful and important tool for functional genomics in plants.Citation1,Citation2 In this technique, CTV-T36, a mild strain of Citrus tristeza virus (CTV), a specific virus of Citrus spp., has been used as a vector to produce dsRNA to silence target genes in citrus plants.Citation3,Citation4 The CTV virus readily multiplies and moves throughout the plant carrying truncated endogenous or exogenous genes. The CTV-based vector has been successfully used to silence phytoene desaturase and delta-aminolaevulinic acid in citrus resulting in specific phenotypes.Citation5,Citation6 In addition, the abnormal wing disc (awd) of the Asian citrus psyllid, Diaphorina citri, the vector of Huanglongbing bacterial pathogen, Candidatus Liberibacter asiaticus was silenced though citrus-mediated RNAi.Citation7
However, the use of the virus-based vector presents some challenges as a consequence of the unequal distribution of the virus within the plant. The unequal distribution of virus results in unpredictable variations in the gene expression and phenotype of the targeted genes. For example, silencing of phytoene desaturase (pds) gene in citrus plants resulted in a variable phenotype, with less gene expression in some branches than others ()), more phenotype in younger leaves than older ()) or the contrast ()), and variations in phenotype between varieties ()).
Figure 1. Photobleaching phenotype is unequally distributed within citrus trees. (a) In Citrus macrophylla (alemow), the phenotype could be more pronounced on one side of the tree than the other side. (b) Within the same branch upper leaves may show more bleaching than the lower leaves, or (c) in some cases phenotype equally appears within the entire branch. (d, e) In Citrus sinensis (L.) Osbeck. (sweet orange) and Citrus aurantium, (sour orange), lower leaves have more photobleaching than upper leaves. (f) Plantlets used to study the influence of the targeted gene sequence orientation in the CTV construct on the RNAi efficiency. (g) Although the virus titers were similar, (h) the reduction of pds expression was significantly more when the anti-sense orientation was used.
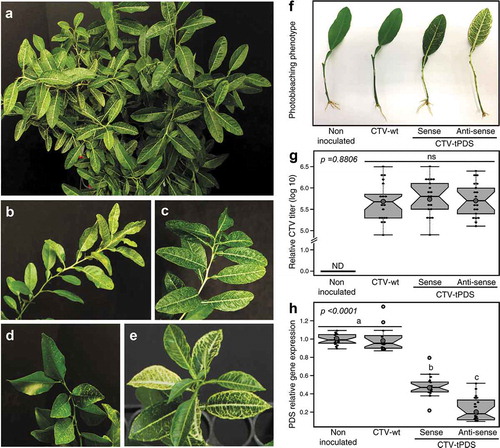
This challenge means that we are unable to fairly compare between treatments such as using the sense or anti-sense orientations of the target gene to construct the CTV vector. In order overcome this problem, we established a simple model by reducing the system from an entire plant to a smaller system consisting of a single leaf, 5 cm of stem and a few roots ()). These plantlets were achieved by dipping the cut leaf stems in rooting hormone and keeping them in a mist bed for two weeks until roots developed. In the current work, the plantlets were made from citrus trees that were previously infiltrated with CTV vector carrying truncated pds in the sense orientation (CTV-tPDS-s) or anti-sense (CTV-tPDS-as) orientation as described in.Citation5 Control plantlets included non-infiltrated trees and wild-type CTV (empty vector). Leaves used to produce the plantlets were selected with pre-phenotypes (photobleaching was not completely pronounced). After rooting (2 wks), we observed that the density of phenotype is higher in plantlets infiltrated with antisense orientation vector, CTV-tPDS-as compared to the sense orientation, CTV-tPDS-s ()). In order to fairly compare the RNA interference efficiency, we demonstrated that there was no significant differences in the virus titers among the treatments ()) while the gene expression levels of pds was significantly lower ()) when anti-sense orientation of pds was used in the CTV construct compared to the sense orientation.
At this point, these observations conclude that the CTV-induced gene silencing is more efficient when the antisense sequence of the target gene is used to construct the vector. In fact, this phenomenon could be explained based on the CTV multiplication with citrus trees.
summarizes the multiplication and the formation of siRNA of CTV-tPDS within citrus plants. The plus-sense RNA genome of CTV acts as mRNA and upon uncoating. It is translated right away using the host cellular machinery to produce the RNA-dependent RNA polymerase (RdRp) (). The RdRp copies the plus-sense genomic RNA into complementary minus-sense RNA. New minus-sense strands serve as the template for new plus-sense genomic strands and subgenomic RNAs synthesis. CTV encodes 10 open reading frames (ORFs) towards the 3ʹ end of the genome, which are expressed by transcription of subgenomic mRNAs (sgRNAs). The RNAi mechanism, when inserts are in both plus-sense and minus-sense orientations (sense and antisense), occurs mainly when the replication intermediate dsRNA is targeted by the host Dicer ()), which generates siRNAs. The siRNAs that are specific to the gene of interest (pds) target the specific mRNA and induce gene silencing phenotype (photobleaching).
Figure 2. Replication of CTV construct in citrus. (a) Genomic map of Citrus tristeza virus (CTV) with the foreign sequence insert position. (b) The replication cycle of CTV as positive single-stranded RNA virus (+ssRNA). (c)The 20 nucleotides siRNAs resulted from the dicer activity and are responsible for the main RNAi of the target gene. (d, e) The subgenomic RNAs (sgRNA) are mostly positive strands (Gowda et al 2001), note the intensity of positive-strand sgRNA compared to negative-strand RNAs. In the case of when the insert is in the antisense orientation, complementary target sequences in the form of sgRNA are produced and enhance the gene silencing effect.
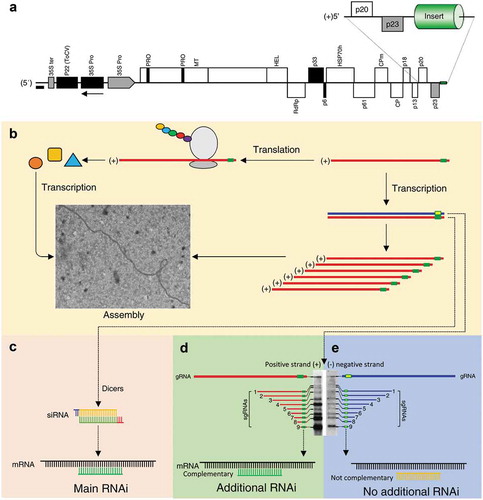
It has been shown that the majority of sgRNA are positive-sense strands.Citation8 When the plus-sense sgRNAs are synthesized by the viral RdRp and the insert is in sense orientation and without complementary target sequence, this sgRNA does not play a role in the gene silencing phenomenon. In the case of when the insert is in the antisense orientation and complementary target sequence, the plus-sense sgRNAs plays a role in the gene silencing phenomenon. With the presence of abundant complementary target sequences in the form of sgRNA, the dsRNAs formed between sgRNA and corresponding mRNA generates more siRNAs and produce additional gene silencing effect compared to the insert in sense orientation ().
In summary, our results showed that the antisense orientation was more efficient than the sense orientation in pds silencing in citrus. Briefly, in the antisense orientation, there is abundant complimentary target sequence (antisense ssRNAs) in the form of sgRNA that bind to the pds mRNAs, and hence prevent their translation. Better understanding of gene silencing mechanisms in higher plants could enhance the use and applications of VIGS technology.
Acknowledgments
The author acknowledges our CREC colleagues for their helpful discussions. I thank Shelley E. Jones for the technical assistance and Lorraine Jones and Floyd Butz for maintaining the trees in greenhouses. This project is funded by the grant no. 201500955-04 from SCRI-NIFA, USDA.
Additional information
Funding
References
- Kumagai MH, Donson J, della-Cioppa G, Harvey D, Hanley K, Grill LK. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc Natl Acad Sci. 1995;92:1679–1683. doi:10.1073/pnas.92.5.1679.
- Ruiz MT, Voinnet O, Baulcombe DC. Initiation and maintenance of virus-induced gene silencing. Plant Cell. 1998;10:937–946. doi:10.1105/tpc.10.6.937.
- Folimonov AS, Bar-Joseph M, Dawson WO. A stable RNA virus-based vector for citrus trees. Virology. 2007;368:205–216. doi:10.1016/j.virol.2007.06.038.
- Dawson WO, Bar-Joseph M, Garnsey SM, Moreno P. Citrus tristeza virus: making an ally from an enemy. Annual review of phytopathology; 2015. Vol. 53. 137–155. doi: 10.1146/annurev-phyto-080614-120012.
- Killiny N, Nehela Y, Hijaz F, Ben-Mahmoud SK, Hajeri S, Gowda S. Citrus tristeza virus-based induced gene silencing of phytoene desaturase is more efficient when antisense orientation is used. Plant Biotechnol Rep. 2019. online first doi:10.1007/s11816-019-00529-0
- Killiny N, Hijaz F, Nehela Y, Hajeri S, Gowda S. Effects of δ-aminolevulinic acid dehydratase silencing on the primary and secondary metabolisms of citrus. Plant Direct. 2018;2:1–19. doi:10.1002/pld3.72.
- Hajeri S, Killiny N, El-Mohtar C, Dawson WO, Gowda S. Citrus tristeza virus-based RNAi in citrus plants induces gene silencing in Diaphorina citri, a phloem-sap sucking insect vector of citrus greening disease (Huanglongbing). J Biotechnol. 2014;176:42–49. doi:10.1016/j.jbiotec.2014.02.010.
- Gowda S, Satyanarayana T, Ayllón M A, Albiach-Martı́ M R, Mawassi M, Rabindran S, Garnsey SM, Dawson WO. Characterization of the cis-acting elements controlling subgenomic mRNAs of Citrus tristeza virus: production of positive- and negative-stranded 3′-terminal and positive-stranded 5′-terminal RNAs. Virology. 2001;286:134–151. doi:10.1006/viro.2001.0987.
