ABSTRACT
Genetic transformation plays a vital role in gene functional study and molecular breeding of soybean. Conventional soybean transformation methods using chemical selectable markers, such as antibiotic or herbicide resistance genes, rely on the identification of positive transgenic lines at advanced developmental stages, making selection procedure labor intensive and time consuming. Utilization of a visual maker to track the transgene would avoid the uncertainty and blindness in the transformation process. In this research, we used green fluorescent protein (GFP) as the selectable marker to detect transgenics at early stages of soybean development. Positive transformants were detected recurrently during each stage of the process based on visualization of the green fluorescence signal, which help us to discard the non-transgenic ones in each stage to reduce the unnecessary experimental cost and lab space. In addition, the positive transgenic seeds can be identified before planting for early detection of transgene and obtain homozygous lines in advance. The method established in this study is also a useful reference for other plant species.
1. Introduction
Molecular biology comes on to a remarkable post-genomic era, the rapid development of sequencing technology triggers data explosion in the biology field. Genomic research has broadening from structural characterization to functional analysis.Citation1 Although genetic mapping, bioinformatics prediction and many other technologies help to draw an inference of gene function, any hypothesis in life science needs a final verification. Genetic transformation depends upon this inference and vice versa, it proves to be a mighty tool in functional genomics. Another significant application of genetic transformation is transgenic breeding. It breaks the species-limits and has been used to increase crop yield, produce biofuels, improve food quality and enhance plant resistance to extreme environments. In addition, the application of genome-editing techniques such as CRISPR/Cas9, also relies on the transformation techniques, which prove the necessity and essentiality of this technology.
Soybean (Glycine max) remains one of the most difficult species to be transformed.Citation2 Currently, the frequently used method is Agrobacterium-mediated transformation, due to the low copy number and greater stability of transgene.Citation3 Previous research has been focusing on how to improve soybean transformation efficiency.Citation4,Citation5 However, there are still existing several other non-negligible problems, including high false-positive transgenic events, complicated detection process of the transgene, and a long transformation cycle to obtain homozygous transgenic lines. Firstly, identification of positive transformants from transgenic events is the dominant part of the technology, especially in species with low transformation efficiency. During tissue culture stages, the majority of regenerated shoots derived from soybean cotyledons are false-positive buds even growing in the medium containing selectable chemicals such as herbicides. Although false-positive transgenic events could be reduced by increasing the herbicide concentration to enhance the selection pressure, which is also detrimental to the regeneration of transgenic explants.Citation6 Secondly, the most common methods to detect transgene include polymerase chain reaction (PCR) tests, herbicide painting, QuickStix Kit for Liberty Link, or β-glucuronidase (GUS) histochemical staining if GUS was used as a reporter gene. All these methods need to collect certain amount of tissues, which is destructive and would affect explant regeneration if early detection of transgenic events is required. In addition, during herbicide resistance screening process, because the transgenic plants have different resistance levels to herbicide, so that researchers cannot make accurate judgments based on the etiolated degrees of leaves, which would result in false-positive problem.Citation7 Thirdly, it normally takes three to four months to obtain T0 transgenic soybean plants. Because of lacking a visual marker to track the transgene, researchers cannot confirm whether the transformation is successful or not until several months later. The uncertainty and blindness in the transformation procedure lead to much more labor, time and materials. However, there are limited researches focusing on above problems.
Green fluorescent protein (GFP) is widely used as a reporter in the field of molecular biology.Citation8 It is convenient to be observed in living cells and tissues at any moment under the fluorescence microscope without any chemical pre-treatment or substrate.Citation9 Therefore, in this study we constructed a plant expression vector containing GFP gene as the only selectable marker instead of GUS or herbicide resistance gene for soybean transformation. We tracked the GFP signal of the calli or regenerated shoots on every explant in each tissue culture stage until obtaining the genetically modified soybean plants. With the aid of GFP signal, we visually tracked the process from Agrobacterium-infected cell forming calli, regenerated shoots, to finally the transgenic soybean plants. Our study confirmed the feasibility of using a visual marker such as GFP for screening and obtaining transgenic plants even for the species recalcitrant to transformation like soybean.
2. Materials and methods
2.1. Plant material, bacterial strain and vector
The soybean variety Williams 82, A. tumefaciens strain EHA105 with rifampicin resistance, and the binary vector pG2RNAi (GenBank accession number: KT954097) which contains a GFP reporter gene directed by the cauliflower mosaic virus (CaMV) promoter were used. The competent EHA105 cells were transformed with pG2RNAi plasmid using heat shock transformation method. The positive clones were screened on LB plates containing 50 mg/L rifampicin and 50 mg/L kanamycin.
2.2. Detection of positive transgenic explants and plants
The explants before transferred to new medium were laid on the petri dish with adaxial side facing downward and the petri dish was sealed with tape. The GFP fluorescence signals were detected using EVOS FL Auto Imaging System (ThermoFisher).
Genomic DNA of four independent transgenic lines and non-transgenic soybean plants was extracted from leaves using the CTAB method.Citation10 The PCR primers for GFP gene were 5ʹ- CCATGTGATCGCGCTTCT −3ʹ (forward) and 5ʹ- GCAAGCTGACCCTGAAGTT’ (reverse). The PCR condition was set as the following: 95°C for 3 min, then 35 cycles of 95°C for 30 s, 60°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 10 min.
2.3. Genetic transformation procedure of soybean
Soybean transformation was carried-out mainly according to the previous published methodCitation5,Citation11 with slight modification (Figure S1 in Supplementary Material):
2.3.1. Agrobacterium solution and explants preparation
Soybean seeds were disinfected with chlorine gas (adding 3.5 ml concentrated hydrochloric acid into 100 ml sodium hypochlorite solution) in a desiccator for 18 h. The EHA105 containing pG2RNAi plasmid was put on a LB plate with 50 mg/L kanamycin and rifampicin to grow for two days, then a single colony was put into 200 ml LB liquid medium growing for 16 h at 28℃, 200 rpm (Figure S1(a) in Supplementary Material). At the same day, all sterilized seeds were soaked into sterilized water for 16–24 h (Figure S1(b) in Supplementary Material).
2.3.2. Explant infection, co-cultivation and GFP detection
Both re-suspension liquid medium and solid co-cultivation medium (CCM) contain 1/10x Gamborg B5 Medium,Citation12 30 g/L sucrose, 3.9 g/L 2[N-morpholino] ethane sulfonic acid (MES) with the pH value of 5.4, except with one difference that solid CCM has 5 g/L agar in it but re-suspension liquid medium does not. The 200 ml Agrobacterium culture was centrifuged and re-suspended with the re-suspension liquid medium to infect soybean half-seeds.
The soaked soybean seeds were excised into two halves along the hilum with a scalpel. Then seed coats were removed and part of the hypocotyledonary axis was sliced off. The obtained explants were put in the infection liquid for at least 30 minutes (Figure S1(c) in Supplementary Material). All infected soybean cotyledons were laid on the sterile filter paper on CCM for 3–5 days, which is the co-culture (CC) stage (Figure S1(d, e) in Supplementary Material). After CC stage, fluorescence detection was performed for all explants before they were transferred to shoot introduction medium (SIM).
2.3.3. Shoot induction and GFP detection
Sterile water containing 50 mg/L cefotaxime and 500 mg/L carbenicillin was used to rinse the explants for three times to remove the excess Agrobacterium outside of the explants. After drying on the clean bench, the cotyledons were inserted into the SIM for shoot induction and differentiation (Figure S1(f) in Supplementary Material). SIM was composed of basal B5 medium with 1.67 mg/L 6-benzylaminopurine, 50 mg/L cefotaxime and 500 mg/L carbenicillin.
All explants were screened for green fluorescence 14 days later before transferred to the second SI stage. The explants showing green fluorescence were marked to distinguish from those that had no fluorescence signal. Both positive and negative transgenic explants were retained and excised brown calli to put on fresh SIM for 14 days (the second SI stage, SI2). At the end of SI2, fluorescence signal detection was performed again on the explants where no fluorescence signal was detected in the first SI stage (SI1). Finally, all explants with fluorescent clustered shoots were retained and transferred to shoot elongation medium (SEM), and negative transgenic explants were removed (Figure S1(g) in Supplementary Material).
2.3.4. Shoot elongation and GFP detection
SEM contained basal MS medium,Citation13 30 g/L sucrose, 0.6 g/L MES, 3 g/L phytagel, 50 mg/L asparagine, 100 mg/L pyroglutamic acid, 0.1 mg/L indole-3-acetic acid, 1 mg/L gibberellic acid, 1 mg/L Zeatin-riboside, 75 mg/L cefotaxime and 500 mg/L carbenicillin and pH value was adjusted to 5.7. The explants with GFP signal were transferred into the SEM. At the SE stage, the elongated shoots with green fluorescence were kept and the non-fluorescent ones were prescind. We change the SEM every two weeks (Figure S1(h) in Supplementary Material).
2.3.5. Rooting, transplanting into soil and growth conditions
The elongated shoots (> 3 cm high) were transferred into the rooting medium (RM), which contained 4.33 g/L salt Mixture (Murashige & Skoog), 20 g/L sucrose, 0.6 g/L MES, 1mg/L Indole-3-butyric acid, 8g/L agar powder and pH was adjusted to 5.7 (Figure S1(i) in Supplementary Material). The plants with many roots (usually took two weeks) were transplanted into pots (soil: vermiculite = 1:1) and grown in growth chamber at 28/25°C with a photoperiod of 16 h/8 h (light/dark) (Figure S1(j) in Supplementary Material).
2.4. Quantitative RT-PCR (qRT-PCR) analysis
Fresh tissues from two transgenic soybean lines were collected for total RNA extraction using Lithium chloride (LiCL) precipitation method.Citation14 Then total RNA was digested using DNase I kit (Cat. No. 18068015, Invitrogen). The first strand cDNA was synthesized using the cDNA Synthesis Kit (Cat. no. 6210A, Takara). The ELF1B (Glyma.02G276600) gene was used as the internal control for relative expression level quantification.Citation15 qPCR reactions were performed using the SYBR Premix Ex Taq kit (Cat. no. RR420A, Takara). The PCR amplification conditions were: 95°C 10 min, 40 cycles of 15 sec at 95°C, and 1 min at 60°C, the melt curve analysis was executed for verifying the specificity of the primer with the following stage: 95°C for 15 sec, 60°C for 1 min, and 95°C for 15 sec. Each sample was repeated three times.
The primer pairs used for qRT-PCR were 5ʹ- CACCTTGATGCCGTTCTTCT −3ʹ (forward) and 5ʹ- GCTGACCCTGAAGTTCATCTG −3ʹ (reverse) for GFP, 5ʹ- CACCTTGATGCCGTTCTTCT −3ʹ (forward) and 5ʹ-GCTGACCCTGAAGTTCATCTG −3ʹ (reverse) for ELF1B.
3. Results
3.1. Detection of transgene in soybean explants based on the green fluorescence signal after co-cultivation stage
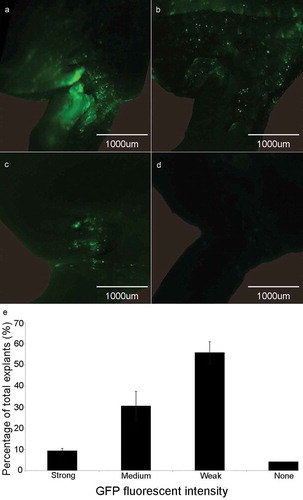
After five days of co-culture with Agrobacterium, soybean half-seed cotyledonary explants were screened for green fluorescence signal using a fluorescence microscope. We detected different strength of green fluorescence signal around the growing points where active growth takes place and form calli. Therefore, the explants were classified into four grades based on GFP signal intensity as follows: almost the entire region around the growing points glowing bright green fluorescence was defined as strong signal ()); medium, some green fluorescence signal but not very bright was defined as medium signal ()); only little detectable fluorescence signal was defined as weak signal ()); and none, without any fluorescence was defined as none signal ()). Among all the Agrobacterium-infected explants, the explants with strong signal accounted for about 9%, the ones with medium signal accounted for 31%, weak signal ones accounted for 56%, and none signal ones accounted for 4% ()). The explants with detectable green fluorescence signals were transferred into SIM for shoot introduction.
Figure 1. GFP signal detection in the cotyledonary explants at co-cultivation stage. (a) Explant with strong fluorescence signal. (b) Explant with medium fluorescence signal. (c) Explant with weak fluorescence signal. (d) Explant with none fluorescence signal. (e) Rate of different fluorescence signal intensity (%) = (the number of explants with strong, medium, weak or none fluorescence signal/total number of explants) × 100%. Results are expressed as mean ± standard error. The experiments were repeated three times with 200 explants for each replicate.
3.2. Screen for positive transformation events based on the green fluorescence signal at shoot introduction and shoot elongation stage
Following the co-cultivation, the explants were cultured in SIM medium for four weeks to induce the formation of calli and shoots (Figure S1(f, g) in Supplementary Material). The green fluorescence signals were not only visible in the explants during CC stage ()), but also detectable in the calli and regenerated shoots during the shoot induction (SI) period (–e)) and shoot elongation (SE) stage (–i)). At the end of the first SI stage (SI1, two weeks after the explants were transferred into SIM), the explants began to form calli, and some of the newly formed calli emitted fluorescence ()). During the second SI stage (SI2), some of the calli differentiated into small buds with green fluorescent signals after about 10 days of transfer to the new SIM ()). At the end of the shoot induction stage (normally four weeks in SIM), some explants showed obvious buds and shoots with green fluorescence (, e)). We tracked the fluorescence signal intensity classification at CC stage () of the explants with positive transgenic shoots at SI2 stage (, e)) and found that 15.87% of the explants with strong fluorescence signal at CC stage had positive transgenic shoots at SI2, and 8.36% of the explants with medium fluorescence signal at CC stage had positive transgenic shoots at SI2 ()). No positive transgenic shoots were observed in the explants with weak signal or none signal ()). On average, 6.94% of the explants had positive transgenic shoots at the end of SI2 stage.
Figure 2. Detection of GFP signal in soybean explants at different stages during tissue culture and transformation efficiency. (a) Co-culture stage. (b) First shoot induction stage. (c) Initial of the second shoot induction stage. (d, e) End of the second shoot induction stage. (f, g, h, i) Shoot elongation stage. (j) Rate of positive transgenic explants at the end of shoot induction stage SI2. Rate of positive transgenic explants at SI2 (%) = (the number of positive transgenic explants at SI2/total number of explants with different GFP signal strength at CC) × 100%. (k) Transformation efficiency after transplanting. Transformation efficiency (%) = (the number of positive plants/total number of explants with different GFP signal strength at CC) × 100%. Results are expressed as mean ± standard error. The experiments were repeated three times with 200 explants for each replicate.
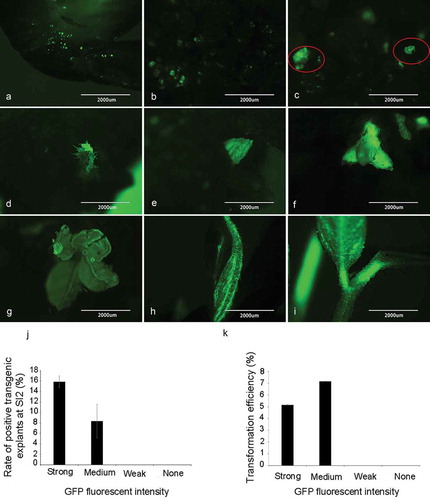
After four weeks in SIM, the positive transgenic explants were all transferred to the SEM and negative ones were all discarded. Before transferring explants into the second SE stage, we detected the green fluorescence signal among the shoots, some of the shoot meristems stems, elongated stems and newly emerged leaves (–h)). We kept the positive buds and cut off the negative ones of explants whose stems have been elongated at the end of the first SE stage. Then the positive explants were cultured for another two weeks in SEM. Then explants with elongated shoots (the height of shoots ≥ 3 cm) having the green fluorescence signal ()) were transferred into the RM.
3.3. Transformation efficiency
We also tracked the fluorescence signal intensity classification at CC stage of the explants that turned into positive transgenic plants in RM, and found that 5.2% of the explants with strong fluorescence signal at CC stage that turned into positive transgenic plants in RM, and 7.16% of the explants with medium fluorescence signal at CC stage that turned into positive transgenic plants in RM, while none positive transgenic plants were obtained from the explants with weak signal or none signal ()). On average, the transformation efficiency was 2.5% ± 0.45%, which means that about 2.5% of the explants got transformed and regenerated into positive transgenic plants (Figure S2 in Supplementary Material).
3.4. Fluorescence signal in T0 transgenic soybean plants
In this study, GFP was under control of the CaMV 35S promoter, a strong constitutive promoter and widely used in different plants.Citation16 We detected the GFP signal in different tissues of the T0 transgenic plants under fluorescence microscope and found the fluorescence signal intensity varies among different tissues (Figure S3 in Supplementary Material): flowers have the strongest GFP signal, followed by leaves and stems, but only very weak fluorescence signal can be detected in roots (Figure S3(a–d) in Supplementary Material). And the GFP expression levels by qRT-PCR analysis (Figure S3(e) in Supplementary Material) were consistent with GFP signal intensity.
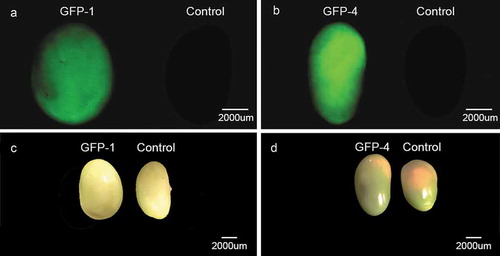
We also detected GFP signals in the seeds of T0 soybean transgenic plants under fluorescence microscope (). Among the T0 transgenic seeds, not all of them have fluorescence signals because of the heterozygous nature of the transgene at this generation. However, by the aid of fluorescence signals of transgene, the positive transgenic seeds can be selected before planting for the next generation, which would accelerate the breeding process and reduce the labor in growing and screening transgenic progenies.
4. Discussion
Since soybean is an important oil crop, an optimized and efficient transformation system for it is necessary. Most plant transformation studies are still based on antibiotic resistance genes to screen transgenic plants, GFP has only been used as a reporter but not selectable marker.Citation17 In this study, we confirmed the feasibility that GFP not only serve as a reporter gene, but also can replace the herbicide resistance gene to screen and select positive transformants, which has the following advantages:
First, positive transformants can be identified dynamically and accurately. GFP fluorescence signal is visible under fluorescence microscope without the need of dyes, which is non-destructive, and therefore can be detected during any stage of the transformation. Since we repeatedly retained the positive transgenic explants based on fluorescence at each tissue cultural stage, the false-negative rate was low for the finally identified transgenic plants.
Second, experimental costs and labor intensity were greatly reduced. Using GFP as the selectable marker and reporter, no herbicide selection is needed to screen the positive transformants, only a fluorescence microscope is enough to identify the transgene, which save a lot of cost on the antibiotics and herbicides. The transformation efficiency in soybean is still low,Citation5,Citation7 which means that most explants during tissue culture are negative transformants. Due to the blindness of traditional transformation method during tissue culture process, both positive and negative transgenic explants were cultured until the end of RM stage, which waste a lot of experimental materials, laboratory space, and labor.
Based on our results, there was very low probability that positive buds occurred from the explants with weak fluorescence. Therefore, in our method, we only kept the explants with strong and medium fluorescence signals and discarded those having weak or no fluorescence signals on which no positive buds would grow. At the end of the second SI stage, we also retained only the positive explants with buds having green fluorescence and transferred them to SEM for stem elongation. During this process, only few explants were retained one month after the explants were infected by Agrobacterium. Finally, only the ones with green fluorescence signals were transferred into RM when their stems were longer than 3 cm. In our new method using GFP as the selectable marker, the experimental cost, lab space and labor are greatly saved compared with the traditional method ()).
Figure 4. Comparison between herbicide and GFP selection-based methods. (a) comparison of the process to screen transgenic events. Left panel shows the process using herbicides and right panel shows the process using GFP signal to screen for positive transgenic events. (b) comparison of the process to select transgenic offspring. Left panel shows selecting homozygous lines by traditional herbicide method. Right panel shows selecting homozygous lines based on GFP signal.
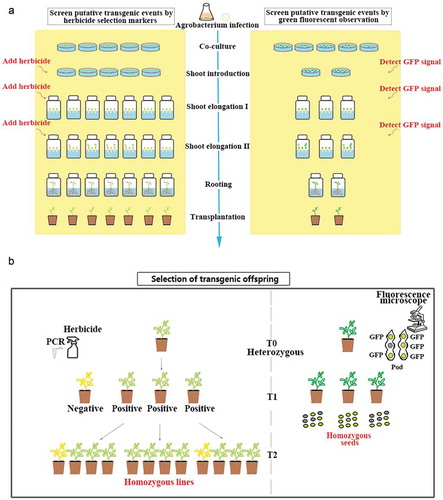
Third, the homozygous lines can be identified at earlier generation. Because the green fluorescence signal can be detected in most tissues of the transgenic soybean plants including seeds, we can select positive transgenic seeds before planting them. We can select the positive transgenic seeds from T0 to T1 generations more easily and rapidly for early detection of transgene and obtain homozygous lines faster than the traditional method, which also save us the time, labor, and space to grow the negative transgenic seeds ()).
We noticed that the regeneration of transgenic cells on antibiotic/herbicide-free media was much higher than antibiotic/herbicide-containing media as previously reported in sunflower.Citation18,Citation19 We also observed that some explants with strong or medium green fluorescence signals would have more than one positive transgenic bud (, f)). However, only limited number of positive explants elongated above 3 cm high. One possible reason is that the SEM we used is suitable for the herbicide screening. Without herbicide selection in this study, the multiple shoots on one explant competed for the nutrients and hormones. Adjusting the ingredient of the SEM may promote more elongated shoots and therefore achieve more positive plants and increase soybean transformation efficiency, which would be further optimized in our future study.
Abbreviations
| CaMV | = | cauliflower mosaic virus |
| CCM | = | co-cultivation medium |
| CC | = | co-culture |
| GFP | = | green fluorescent protein |
| PCR | = | polymerase chain reaction |
| RM | = | rooting medium |
| SEM | = | shoot elongation medium |
| SIM | = | shoot introduction medium |
Supplemental Material
Download Zip (5.2 MB)Acknowledgments
The authors thank Sujata Agarwal at University of Tennessee for her help on using the experimental instruments.
Disclosure statement
No potential conflict of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Medini D, Serruto D, Parkhill J, Relman DA, Donati C, Moxon R, Falkow S, Rappuoli R. Microbiology in the post-genomic era. Nat Rev Microbiol. 2008;6:419–430. doi:10.1038/nrmicro1901.
- Zhang YM, Liu ZH, Yang RJ, Li GL, Guo XL, Zhang HN, Zhang HM, Di R, Zhao QS, Zhang MC. Improvement of soybean transformation via Agrobacterium tumefaciens methods involving alpha-aminooxyacetic acid and sonication treatments enlightened by gene expression profile analysis. Plant Cell Rep. 2016;35:1259–1271. doi:10.1007/s00299-016-1958-2.
- Travella S, Ross SM, Harden J, Everett C, Snape JW, Harwood WA. A comparison of transgenic barley lines produced by particle bombardment and Agrobacterium-mediated techniques. Plant Cell Rep. 2005;23:780–789. doi:10.1007/s00299-004-0892-x.
- Jia YY, Yao XD, Zhao MZ, Zhao Q, Du YL, Yu CM, Xie FT. Comparison of soybean transformation efficiency and plant factors affecting transformation during the Agrobacterium infection process. Int J Mol Sci. 2015;16:18522–18543. doi: 10.3390/ijms160818522.
- Li SX, Cong YH, Liu YP, Wang TT, Shuai Q, Chen NN, Gai JY, Li Y. Optimization of Agrobacterium-mediated transformation in soybean. Front Plant Sci. 2017;8:246. doi: 10.3389/fpls.2017.00246.
- Azhakanandam K, Silverstone A, Daniell H, Davey MR. Recent advancements in gene expression and enabling technologies in crop plants. New York: Springer Verlag publishers. 2015.
- Song ZY, Tian JL, Fu WZ, Li L, Lu LH, Zhou L, Shan ZH, Tang GX, Shou HX. Screening Chinese soybean genotypes for Agrobacterium-mediated genetic transformation suitability. J Zhejiang Univ-Sc B. 2013;14:289–298. doi:10.1631/jzus.B1200278.
- Leffel SM, Mabon SA, Stewart CN. Applications of green fluorescent protein in plants. Biotechniques. 1997;23:912. doi:10.2144/97235bi01.
- de Ruijter NCA, Verhees J, van Leeuwen W, van der Krol AR. Evaluation and comparison of the GUS, LUC and GFP reporter system for gene expression studies in plants. Plant Biol. 2003;5:103–115. doi:10.1055/s-2003-40722.
- Paterson AH, Brubaker CL, Wendel JF. A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol Biol Rep. 1993;11:122–127. doi:10.1007/BF02670470.
- Paz MM, Martinez JC, Kalvig AB, Fonger TM, Wang K. Improved cotyledonary node method using an alternative explant derived from mature seed for efficient Agrobacterium-mediated soybean transformation. Plant Cell Rep. 2006;25:206–213. doi:10.1007/s00299-005-0048-7.
- Gamborg OL, Miller RA, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–158. doi:10.1016/0014-4827(68)90403-5.
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plantarum. 1962;15:473–497. doi:10.1111/j.1399-3054.1962.tb08052.x.
- Cathala G, Savouret JF, Mendez B, West BL, Karin M, Martial JA, Baxter JD. A method for isolation of intact, translationally active ribonucleic acid. DNA. 1983;2:329–335. doi:10.1089/dna.1983.2.329.
- Wan Q, Chen SL, Shan ZH, Yang ZL, Chen LM, Zhang CJ, Yuan SL, Hao QN, Zhang XJ, Qiu DZ, et al. Stability evaluation of reference genes for gene expression analysis by RT-qPCR in soybean under different conditions. PLoS One. 2017;12:e0189405. doi:10.1371/journal.pone.0189405.
- Kay R, Chan A, Daly M, Mcpherson J. Duplication of CaMV35S promoter sequences creates a strong enhancer for plant genes. Science. 1987;236:1299–1302. doi:10.1126/science.236.4806.1299.
- Ahlandsberg S, Sathish P, Sun CX, Jansson C. Green fluorescent protein as a reporter system in the transformation of barley cultivars. Physiol Plantarum. 1999;107:194–200. doi:10.1034/j.1399-3054.1999.100207.x.
- Hewezi T, Perrault A, Alibert G, Kallerhoff J. Dehydrating immature embryo split apices and rehydrating with Agrobacterium tumefaciens: A new method for genetically transforming recalcitrant sunflower. Plant Mol Biol Rep. 2002;20:335–345. doi:10.1007/BF02772121.
- Hewezi T, Jardinaud F, Alibert G, Kallerhoff J. A new approach for efficient regeneration of a recalcitrant genotype of sunflower (Helianthus annuus) by organogenesis induction on split embryonic. Plant Cell Tissue Organ Cult. 2003;73:81–86. doi:10.1023/A:1022689229547.