ABSTRACT
Nitrogen (N) is an essential macronutrient for optimal plant growth and ultimately for crop productivity Nitrate serves as the main N source for most plants. Although it seems a well-established fact that nitrate concentration affects flowering, its molecular mode of action in flowering time regulation was poorly understood. We recently found how nitrate, present at the shoot apical meristem (SAM), controls flowering time In this short communication, we present data on the tissue-specific expression patterns of NITRATE REDUCTASE 1 (NIA1) and NIA2 in planta. We show that transcripts of both genes are present throughout the life cycle of Arabidopsis thaliana plants with NIA1 being predominantly active in leaves and NIA2 in meristematic tissues.
Introduction
In Arabidopsis thaliana (Arabidopsis), nitrate, once taken up by roots, is distributed to sink tissues including meristems via xylem vessels.Citation1 It is assimilated into ammonia by nitrate reductase, encoded by NITRATE REDUCTASE 1 (NIA1) and NIA2, and further converted into amino acids,Citation2 which serve to support biological processes. Our previous analyses showed that nitrate is present at the shoot apical meristem (SAM) of Arabidopsis plants, where it directly triggers flowering.Citation3 We demonstrated that a moderate, non-stressful restriction of nitrate availability delays flowering time, due to decreased expression of SUPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 (SPL3) and SPL5 at the SAM. Moreover, we found that the nitrate-inducible genes NIA1 and NIA2 and the master regulators of nitrate signaling, NIN-LIKE PROTEIN 6 (NLP6) and NLP7, Citation4 are present at the SAM. We concluded that nitrate controls the flowering time at the SAM through NLP6 and NLP7 by regulating SOC1 at least in part via SPL3 and SPL5.Citation3
Interestingly, we found that NIA1 and NIA2 expression patterns at the SAM were barely overlapping with each other, suggesting that they might be regulated independently and assimilate nitrate in a stage- and tissue-dependent manner. Since their transcriptional response rapidly and directly responds to nitrate and does not require de novo protein synthesis, Citation5 the analysis of NIA transcripts provides information on the status of nitrate signaling and assimilation in cells. Although the biological function of NIA1 and NIA2 [e.g. Citation6] and nitrate-dependent expression of promoter-reporter constructsCitation7,Citation8 were previously studied, we provide first tissue- and cell-specific expression analyses for the first step of nitrate assimilation in planta.
Results
We made use of RNA in situ hybridizations, in order to investigate cell- and tissue-specific expression of NIA1 and NIA2 transcripts in planta. Using tissue-specific probes for NIA1 and NIA2,Citation3 we detected transcripts in various tissues of Arabidopsis wild-type plants. Representative images of different tissue sections are presented in and .
Figure 1. NIA1 expression in Arabidopsis thaliana plants. RNA in situ hybridization using a specific probe for Arabidopsis NIA1 on transversal sections through the hypocotyl (A), longitudinal sections through a root tip (B), rosette leaves (C), the vegetative shoot apical meristem (D), the inflorescence meristem (E) and young pistils, containing ovules (F-G). Representative stains in developing embryos (H-I): early (H) and late heart stages (I). Inset in H: globular stage embryo. Scale bars 20µm (A-D, F-I) and 50µm (E).
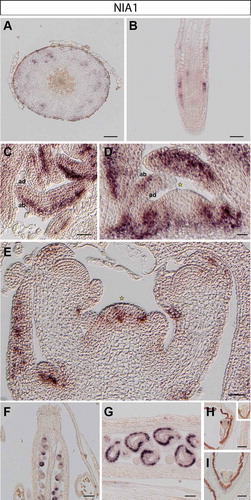
Figure 2. NIA2 expression in Arabidopsis thaliana plants. RNA in situ hybridization using a specific probe for Arabidopsis NIA2 on transversal sections through the hypocotyl (A), longitudinal sections through an emerging lateral root tip (B), rosette leaves (C), the vegetative shoot apical meristem (D), the inflorescence meristem (E) and various stages of pistil development (F-G). Inset in E: replum and upper part of pedicel. Representative stains in developing embryos (H-I): early (H) and late heart stages (I). Inset in H: globular stage embryo. Scale bars 20µm (A-D, F-I) and 50µm (E).
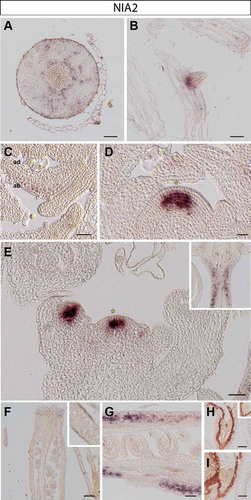
In the hypocotyl, NIA1 is present in distal phloem cells but absent from proximal tissue including phloem, cambium and xylem cells ()). In contrast, NIA2 is expressed throughout the cambium (likely including the bifacial stem cells and distal cambium) and phloem cells throughout a cross section of the hypocotyl ()).
In roots, NIA1 is expressed in epidermis cells of the root meristem, with some weaker expression in vascular layers, presumably in phloem cells ()). We did not observe any NIA2 expression in main root meristematic zones (not shown); however, transcript of NIA2 is found at the base of newly emerging lateral root primordia, coinciding with procambium and young vasculature cellsCitation9 ()).
We found that NIA1 is predominantly present in all leaf stages, including newly formed leaf primordia, spreading from the proximal to the distal part of young leaves. Its expression domain is, however, restricted to the abaxial side, which corresponds to spongy mesophyll cellsCitation10 (). Instead, transcript of NIA2 is barely detectable in leaves and fully absent from young leaf primordia ().
NIA1 transcript is also present in cauline leaves of the inflorescence apex ()). Moreover, as previously observed,Citation3 NIA1 is expressed only in the rib and peripheral zones of the SAM and axillary meristems, but is not present in the central region including the organizing center and stem cell niche of the vegetative ()) and the inflorescence SAM ()). NIA2 is strongly expressed in the center of the vegetative ()), inflorescence and floral meristems ()).Citation3 Starting in L3 the NIA2 expression domain overlaps with the organizing center of the SAM and the region lacking NIA1 transcript (compare ) and )).
As different expression patterns of NIAs are observed in floral meristems, we also analyzed their expression in crucial stages of seed development. Expression of NIA1 is found in the cells of the inner integument in ovules (), where NIA2 is not detectable (). In contrast, the signal of NIA2 is found in valves of siliques ()).
After fertilization, NIA1 is expressed in the various stages of embryo development. We found that NIA1 expression is more abundant at the abaxial side of newly formed cotyledons in early heart stage embryosCitation11 ()). In later heart stage embryos, expression of NIA1 is present in protodermal cells in the basal portion of the embryo,Citation12 in the region destined to give rise to the hypocotyl (). NIA1 transcript is not detectable in globular (inset of )) or mature embryos (not shown). In contrast, NIA2 is expressed in procambial cells in the central zone of early and later heart stagesCitation13 (), but is absent from the globular (inset of )) and more mature stages of embryogenesis (data not shown).
Conclusions
We found that the expression pattern of NIA1 complemented that of NIA2 in the same organ within the set of tissue samples analyzed, suggesting that nitrate is assimilated by nitrate reductase encoded by either NIA1 or NIA2 in a cell- or tissue-specific manner. This further indicates non-redundant functions of the two NIAs in nitrate assimilation that might be associated with cell specification and plant development. This is in agreement with previous reports showing that single mutants of nia1 and nia2 showed different responses and sensitivity levels to, e.g. salicylic acidCitation14 or cytokinin treatments.Citation15 It should, however, be kept in mind that expression of both genes is regulated by various other endogenous and exogenous factors resulting in a diurnally differential expression pattern, Citation16 which might result in changes of expression domains not covered in this study.
NIA1 function seems to be predominantly restricted to leaf tissues. Apart from rosette leaves, this includes cotyledon primordia, cauline leaves, and cells within organ primordia destined to become leaf-like organs, but also the inner integument of developing ovules. The establishment of the inner ovule layer is known to be the earliest morphological sign of the adaxial-abaxial polarity.Citation17 The localization of NIA1 transcript at the abaxial side of newly formed cotyledons during embryogenesis, rosette leaves, and the inner layer of the integument suggests that NIA1 might be involved in the spatiotemporal regulation of polarity in general. NIA2, on the other hand, is active in highly dividing meristematic regions such as within the SAM and cambial cells. The analysis of the single and double nia knockout mutants would be required to elucidate the role of NIA1 in the establishment of the abaxial-adaxial polarity and NIA2 in the regulation of meristematic niches in plants.
In summary, our results suggest that both NIAs take part in other biological and developmental processes apart from their well-established metabolic function.
Materials and methods
Plants material and growth conditions
Arabidopsis thaliana accession Columbia (Col-0) was used in this study. Plants were grown in controlled growth chambers (Percival Scientific Inc., USA) or in the greenhouse at 22°C in long- (16-h light/8-h dark) or short-day (8-h light/16-h dark) photoperiods. Plant material for embedding was generally harvested toward the end of the day.
RNA in situ hybridization
For RNA in situ hybridization apex, hypocotyl, root, silique and flower samples were harvested, fixed with FAA solution (formaldehyde, ethanol, acetic acid; ASP300S, Leica) and embedded into wax (EG1160, Leica) as previously reported.Citation18 RNA in situ hybridization was carried out as described.Citation18 Probes for NIA1 and NIA2 were previously published.Citation3
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Christin Abel and Tanja Seibert for tissue sectioning and performing some of the hybridization assays. Work in the Wahl group is supported by the BMBF (031B0191), DFG grants within the SPP1530 (WA3639/1-2, 2-1) and the Max-Planck-Society. J.J.O. thanks the DFG for funding the Collaborative Research Center 973.
Additional information
Funding
References
- Dechorgnat J, Nguyen CT, Armengaud P, Jossier M, Diatloff E, Filleur S, Daniel-Vedele F. From the soil to the seeds: the long journey of nitrate in plants. J Exp Bot. 2011;62(4):1349–1359. doi:10.1093/jxb/erq409.
- Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A. Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot. 2010;105(7):1141–1157. doi:10.1093/aob/mcq028.
- Olas JJ, Van Dingenen J, Abel C, Dzialo MA, Feil R, Krapp A, Schlereth A, Wahl V. Nitrate acts at the Arabidopsis thaliana shoot apical meristem to regulate flowering time. New Phytol. 2019;223(2):814–827. doi:10.1111/nph.15812.
- Marchive C, Roudier F, Castaings L, Brehaut V, Blondet E, Colot V, Meyer C, Krapp A. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat Commun. 2013;4:1713. doi:10.1038/ncomms2650.
- Gowri G, Kenis JD, Ingemarsson B, Redinbaugh MG, Campbell WH. Nitrate reductase transcript is expressed in the primary response of maize to environmental nitrate. Plant Mol Biol. 1992;18:55–64.
- Wilkinson JQ, Crawford NM. Identification and characterization of a chlorate-resistant mutant of Arabidopsis thaliana with mutations in both nitrate reductase structural genes NIA1 and NIA2. Mol Gen Genet. 1993;239(1–2):289–297. doi:10.1007/bf00281630.
- Yang H, Zhou Y, Zhang Y, Wang J, Shi H. Identification of transcription factors of nitrate reductase gene promoters and NRE2 cis-element through yeast one-hybrid screening in Nicotiana tabacum. BMC Plant Biol. 2019;19(1):145. doi:10.1186/s12870-019-1724-z.
- Konishi M, Yanagisawa S. Roles of the transcriptional regulation mediated by the nitrate-responsive cis-element in higher plants. Biochem Biophys Res Commun. 2011;411(4):708–713. doi:10.1016/j.bbrc.2011.07.008.
- Porco S, Larrieu A, Du Y, Gaudinier A, Goh T, Swarup K, Swarup R, Kuempers B, Bishopp A, Lavenus J, et al. Lateral root emergence in Arabidopsis is dependent on transcription factor LBD29 regulation of auxin influx carrier LAX3. Development. 2016;143(18):3340–3349. doi:10.1242/dev.136283.
- Chitwood DH, Guo M, Nogueira FT, Timmermans MC. Establishing leaf polarity: the role of small RNAs and positional signals in the shoot apex. Development. 2007;134(5):813–823. doi:10.1242/dev.000497.
- Chandler JW. Cotyledon organogenesis. J Exp Bot. 2008;59(11):2917–2931. doi:10.1093/jxb/ern167.
- Lin Y, Schiefelbein J. Embryonic control of epidermal cell patterning in the root and hypocotyl of Arabidopsis. Development. 2001;128:3697–3705.
- Jouannet V, Brackmann K, Greb T. (Pro)cambium formation and proliferation: two sides of the same coin? Curr Opin Plant Biol. 2015;23:54–60. doi:10.1016/j.pbi.2014.10.010.
- Hao F, Zhao S, Dong H, Zhang H, Sun L, Miao C. Nia1 and Nia2 are involved in exogenous salicylic acid-induced nitric oxide generation and stomatal closure in Arabidopsis. J Integr Plant Biol. 2010;52(3):298–307. doi:10.1111/j.1744-7909.2010.00920.x.
- Yu X, Sukumaran S, Mrton L. Differential expression of the arabidopsis nia1 and nia2 genes. cytokinin-induced nitrate reductase activity is correlated with increased nia1 transcription and mrna levels. Plant Physiol. 1998;116(3):1091–1096. doi:10.1104/pp.116.3.1091.
- Galangau F, Daniel-Vedele F, Moureaux T, Dorbe MF, Leydecker MT, Caboche M. Expression of leaf nitrate reductase genes from tomato and tobacco in relation to light-dark regimes and nitrate supply. Plant Physiol. 1988;88(2):383–388. doi:10.1104/pp.88.2.383.
- Sieber P, Gheyselinck J, Gross-Hardt R, Laux T, Grossniklaus U, Schneitz K. Pattern formation during early ovule development in Arabidopsis thaliana. Dev Biol. 2004;273(2):321–334. doi:10.1016/j.ydbio.2004.05.037.
- Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science. 2013;339(6120):704–707. doi:10.1126/science.1230406.
