ABSTRACT
Prohibitins (PHBs) are composed of an obviously conserved protein family in eukaryotic cells. Despite the extensive and in-depth research of mammalian PHB1 and PHB2, the plant prohibitins functions are not completely elucidated and little is known about their mechanism of action. This review focuses on the current knowledge about the protein subcellular localization, interaction proteins and target genes of PHB3. Furthermore, we discussed the roles of PHB3 protein in plant growth and development, plant responses to abiotic or biotic stresses and its participation in phytohormonal signaling.
KEYWORDS:
Prohibitins (PHBs) are composed of an obviously conserved protein family in eukaryotic cells and primitively discovered as tumor suppressor factor in mammalian cells.Citation1 Prohibitins play an essential role in a series of cellular processes, including mitochondrial respiration,Citation2,Citation3 cell cycle regulation,Citation4,Citation5 receptor-mediated signaling at the cell surface,Citation6 cell deathCitation7 and aging.Citation8 Prohibitins obviously function diversity also reflected in its various subcellular location, including mitochondria,Citation8,Citation9 nuclearCitation4,Citation7 and plasma membrane.Citation6,Citation10 Phylogenetic analysis showed that prohibitins in all species can be classified into two types depend on the phylogenetic relationships with yeast PHB1 and PHB2. In Arabidopsis, prohibitins were divided into type-I PHB3 and PHB4, type-II PHB1, PHB2 and PHB6. All prohibitin genes are chiefly expressed in both shoot and root actively proliferative tissues and dividing cells in Arabidopsis.Citation11–Citation13 Previous study showed that plant prohibitins are needed to play a crucial role in cell division, senescence, and root hair elongatio,Citation11,Citation12,Citation14,Citation15 as well as response to ethylene signaling,Citation16 oxidative stress,Citation11 plant defense,Citation17 nitric oxide (NO) accumulation.Citation18 While plant prohibitins have been implicated in the regulation of plant development, there is currently no more information regarding the exact role and function mechanism of plant prohibitins.
In Arabidopsis, PHB3 and PHB6 have the highest homology with mammalian PHB1 and PHB2, respectively.Citation12 Mammalian PHB1 and PHB2 can form complex to regulate a series cellular and developmental processes. By far, almost all of the researches on plant prohibitin genes are focusing on PHB3, as the PHB3 knock-out mutant has pleiotropic phenotypes. All of the research results showed that PHB3 would be a key plant developmental regulator and PHB3 has the conserved function as mammalian PHB1 and PHB2. Thus, the present review looks back the latest progress on the localization, functions, interacting proteins and target genes of PHB3, to provide ideas for the further research on PHB3.
The localization of the PHB3
Subcellular localization of PHB3 in the leaves of N. benthamiana indicates that it is found in several cellular locations, including the nucleus and throughout the cytoplasm.Citation16 No canonical mitochondrial transit peptide or nuclear localization signals were detected in the PHB3 protein sequences, but the N-terminal peptide was necessary for targeting to the mitochondria.Citation12 The PHB3 mitochondrial localization was confirmed in subsequently studies.Citation19,Citation20 Intriguingly, PHB3 also was localized in the nucleus.Citation16,Citation20 The PHB3 protein with the nuclear localization signal (NLS) but not with the nuclear export signal (NES) in the N-terminal can rescue the phb3 developmental deficiency completely.Citation20 Moreover, PHB3 also localizes to chloroplasts using fractionation, protease protection, and live imaging.Citation21 Except of the above localization, it also was found located in the Golgi and endoplasmic reticulum. In the Arabidopsis protoplast, the PHB3 can co-localization with the Golgi marker and endoplasmic reticulum marker ().
Figure 1. The Golgi and endoplasmic reticulum localization of PHB3. A, PHB3-YFP and endoplasmic reticulum marker CNX-RFP are expressed in the indicated protoplasts. The CNX-RFP signal (blue), PHB3-YFP signal (yellow), and merged photos are shown from representative samples. The red arrows indicate the co-localization in endoplasmic reticulum. Scale bar = 20 μm. B, PHB3-YFP and Golgi marker Man1-RFP are expressed in the indicated protoplasts. The Man1-RFP signal (blue), PHB3-YFP signal (yellow), and merged photos are shown from representative samples. The red arrows indicate the co-localization in Golgi. Scale bar = 20 μm.
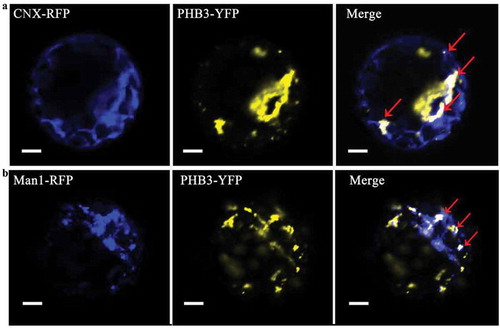
The PHB3 localization diversity decided its pleiotropic roles in plant development. Though there is no predicted mitochondrial signal peptide, no predicted transmembrane domain, or nuclear localization signal in the PHB3 amino acid sequence (data not shown), the putative localization signal peptide sequence of the PHB3 is needed more protein truncation and amino acid mutation experiments to detect. Otherwise, PHB3 may localize to different organelles through protein-protein interaction. Thus, the PHB3 interaction proteins will be very important for its subcellular localization and function. The KDEL receptor (KDELR) is a seven-transmembrane-domain protein involved in retrograde transport of protein chaperones from the Golgi complex to the endoplasmic reticulum. Giannotta et al. (2015) found that mammalian prohibitin-1 (PHB) act as a KDELR interactor and absence of PHB decreases the levels of the Golgi-localised KDELR and prevents KDELR-dependent Golgi-to-plasma-membrane transport. These mysterious issues are very intriguing and need more refined experiments to uncover.
The interacting proteins of the PHB3
It has been shown that PHB3 protein co-purifies with other PHB proteins (PHB1, PHB2, PHB4, and PHB6), presumably forming with them a hetero-oligomeric high molecular weight complexes in mitochondria.Citation12 Moreover, complexes (1 MDa) formed of prohibitins have been identified in the mitochondria of plants.Citation22 Similar to that, PHB3 has been found to form complex with PHB6 using the new protein interaction technology, though their genetic interaction mechanism and the complex function are unclear.Citation23 In plants, prohibitins closely interact with m-AAA proteases and together form 2 MDa m-AAA-PHB complexes.Citation22 Intriguingly, Arabidopsis PHB3 has been found to interact with the enzyme ISOCHORISMATE SYNTHASE1 (ICS1) when analyzed proteins that coimmunoprecipitated with ICS1 via mass spectrometry.Citation21 Recently, PHB3 has been co-immunoprecipitated with the nuclear envelope and mitochondria localized RNA-binding protein OPENER.Citation24
In the mammalian, PHB has been verified to interact with chromatin remodeling factors to regulate gene expression. PHB has been reported to repress E2F via recruitment of the repressive proteins histone deacetylase 1 (HDAC1), nuclear receptor co-repressor (N-CoR) and the chromatin-condensing proteins brahma-related gene-1 (BRG1)/brahma (Brm).Citation4,Citation25 However, the same mechanism has not found in plants yet.Citation20,Citation26 Arabidopsis PHB3 was mentioned to be interacted strongly in the yeast two-hybrid assay with a component of the TAFIID complex, Taf12b.Citation16 But the experiment data has not been shown yet. It will be worthy to test that which kind of the gene transcription regulators, e.g., histone acetylases and deacetylases, may be the PHB3 interaction protein.
The function of the PHB3
Previous studies showed that PHB3 is prominently expressed in meristem tissues and phb3 null mutants exhibit meristematic cell proliferation defects.Citation12,Citation13,Citation19,Citation20,Citation22 It was found that loss of function of the homologous PHB3 caused mitochondrial swelling, decreased meristematic cell production, increased cell division time and reduced cell expansion rates, leading to severe growth retardation. Very interesting, transgenic lines overexpressing PHB3 also showed leaf shape aberrations and an increased shoot branching phenotype.Citation12 These results indicate that PHB3 homeostasis in the plant is critical for the plant growth and development. PHB3 was recognized to be necessary for proficient mitochondrial function or biogenesis to maintain cell division and differentiation in apical tissues.Citation12 In order to supporting this conclusion, the PHB3 interacting protein OPENER in the mitochondria also has similar mitochondrial size, meristem expression and root meristem defects.Citation23 Though the PHB3 play important roles in meristematic cell proliferation, the exact regulating mechanism is not clear until this year. PHB3 control the cell proliferation partially through regulating the MCM genes expression level to maintain the genomic stability and cell cycle in the nucleus.Citation20 Further more, PHB3 maintains root stem cell niche identity and cell division partially through ROS-responsive AP2/ERF transcription factors such as ERF109, ERF114, and ERF115.Citation19 Thus, PHB3 was thought to be one of the retrograde signaling resources from mitochondria to nucleus.Citation26 In spite of the PHB3 mutation can cause significant reactive oxygen species (ROS) accumulation, the retrograde signaling may tranduce into nucleus through the ROS-dependent and ROS-independent ways.Citation19,Citation20,Citation27
Phytohormone is very important for the plant development. PHB3 loss-of-function mutant was identified by screening for Arabidopsis mutant seedlings with an enhanced ethylene response, and PHB3 functions downstream of EIN2 as a positive regulator of expression of a subset of ethylene-regulated genes along with a group of genes required for maintaining growth in the presence of ethylene.Citation16 Moreover, phb3 mutant was found be defective in auxin-induced lateral root formation,Citation18 and auxin accumulation, transport as well as signaling.Citation19,Citation20 The PHB3 null allele mutants and PHB3 over-expressions all showed leaf shape aberrations and abnormal plant architechture.Citation12 Virus-induced gene silencing of PHBs in Petunia hybrida caused abnormal leaf morphology.Citation14 These phenotypes may be resulted from the decreased auxin content or defected auxin signaling as the mitochondrial localized FtSH4 mutant.Citation20,Citation28 PHB3 is necessary for the leaf longevity because PHB3 mutation resulted in accelerated senescence.Citation12,Citation14 Similarly, mitochondrial FtSH4 also take participate in leaf senescence through SA content and SA signaling.Citation29 Intriguingly, PHB3 also regulates SA biosynthesis by forming PHB3-ICS1 complexes to stabilize ICS1 and promote SA production in response to stress.Citation21 Thus, it can be reasonable to predict that PHB3 also regulates leaf senescence through SA. SA is necessary for the plant cell death and autophagy,Citation29 and the mammalian inner mitochondrialmembrane protein PHB2 was identified as a crucial mitophagy receptor involved in targeting mitochondria for autophagic degradation.Citation30 It will be a very interesting point to explore the relationship between Arabidopsis PHB3 and autophagy or mitophagy.
Prohibitin accumulation was found to be a common cellular response to different stressing stimuli including the ER stress, oxidative phosphorylation stress, mitochondrial organization and homeostasis stress, as well as the unfolded protein response (UPR).Citation31 Plant PHB3 also was found to be response to all kinds of stresses. The point mutant phb3 that converting the highly conserved Gly-37 to an Asp in the protein’s SPFH domain and the knock-out phb3 mutant were defective in H2O2-induced NO accumulation, abscisic acid-induced NO accumulation and stomatal closure. Both mutants were less sensitive to salt stress, showing no increase in NO accumulation and less inhibition of primary root growth in response to NaCl treatment. In addition, light-induced NO accumulation was dramatically reduced in cotyledons.Citation18 Recently, the phb3 mutant displayed reduced levels of SA, the SA-regulated protein PR1, and hypersensitive cell death in response to UV-C and avirulent strains of Pseudomonas syringae.Citation21 And phb3 mutant was sensitive to DNA damage agent and oxidation stress.Citation20
The target genes of the PHB3
By far, the verified PHB3 target gene is largely unclear. Using the microarray, the down- and up-regulated genes in the phb3 mutants were associated with mitochondrial and stress-related transcripts.Citation12 The expressions of a subset of ethylene-regulated genes were affected in the phb3 mutant.Citation16 However, whether those genes are PHB3 direct or indirect target genes are waiting for certification by experiments. The only identified direct targets are the minichromosome maintenance (MCM) genes, especially one member of the MCM2. PHB3 acts as transcriptional co-regulator to repress the expression of MCM2 and other MCM members by binding to the promoter E2F–cis-acting elements.Citation20 Moreover, the retrograde signalling in mutants defective in mitochondrial protein prohibitin phb3 can be suppressed by knocking out the transcription factor ANAC017.Citation26 Whether the PHB3 can bind on the promoter and regulate the expression of ANAC017 is deserved to be analyzed. Intriguingly, a putative PHB3-binding site with ATTCCCGG sequence, which conform to the Arabidopsis E2Fa specific binding site with a WTTSSCSS (where W = A or T and S = G or C) cis-acting consensus element,Citation32 was found in the 5ʹ UTR region of ANAC017. PHB3 maintains root stem cell niche identity through ROS-responsive AP2/ERF transcription factors such as ERF109, ERF114, and ERF115, however, the repressing mechanism of PHB3 on ERF109, ERF114 or ERF115 transcript is unclear.Citation19 There are none E2Fa specific binding cis-acting consensus element in the promoter regions of ERF109, ERF114 and ERF115 genes. Thus, PHB3 may regulate a series of genes such as ERF genes as an indirect way. Therefore, the whole genomic ChIP-Sequence is needed to perform in the upcoming research to discover the putative PHB3 binding sites and to search for the direct target genes of PHB3.
Conclusion
In conclusion, it is proposed that PHB3 function as negative or positive co-regulator of transcription to exert influence on cell cycle and cell proliferation and to regulate the expression of series target genes in regulating plant development in Arabidopsis. It is possible that the PHB3 may be induced and translocated into different subcellulars such as mitochondria, nucleus, chloroplasts, Golgi and endoplasmic reticulum to regulate the phytohormone signaling, genome stability and cell proliferation when the plant encounters the biotic or abiotic stress or the cell in a certain condition such as DNA damage agent and oxidation stress. In the mitochondria, chloroplasts and other cytoplasmic organelles, PHB3 formed different protein complex to regulate the mitochondrial integrity and redox equilibrium. Upon PHB3 loss-of-function, the ROS and the different phytohormones such as SA, ethylene, and auxin contents and signaling will be out of homeostasis. Subsequently, different retrograde signaling caused by the PHB3 transduced into the nucleus. In the nucleus, PHB3 binds directly on the target genes, e.g. MCM2; or regulates indirectly on the ROS-responsive genes, e.g. ERF115, ERF114 and ERF109, to regulate the cell cycle and cell proliferation. Upon PHB3 loss-of-function, its repressed genes or promoted will be elevated or decreased out of control and result in cell cycle arrest. Consequently, the stem cell niche modulators are repressed or altered to prevent meristem cell from proliferation and the plant growth and development are changed. Since the function mechanism difference between plant and mammalian nuclear prohibitin, some processes and important questions involved in plant PHB3 need to be identified. Firstly, what are the PHB3 function in different organelles, especially in the Golgi and ER? Secondly, how does the PHB3 translocate between the cytoplasm and nucleus? Thirdly, how is the biotic and abiotic stress sensed by and transducted to the PHB3? Whether the protein kinases regulate PHB3 to response the stress and development signaling? Finally, whether does the PHB3 regulate other E2F-cis-acting elements containing and no containing genes directly? Hence, the targets and partners discovery for PHB3 will provide more evidence to expound its activities in the nucleus and the communication between cytoplasm and nucleus. In further, the detail regulation mechanisms for PHB3 regulation on cell proliferation, phytohormone signaling, responses to abiotic and biotic stress need more evidences to elucidate.
Acknowlegement
This work is supported by the National Natural Science Foundation of China (31870301, 31370350), and Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2016 for S. Z.).
Additional information
Funding
References
- Mcclung J, Danner DB, Stewart DA, Smith JR, Schneider EL, Lumpkin CK, Dell’Orco RT, Nuell MJ. Isolation of a cDNA that hybrid selects antiproliferative mRNA from rat liver. Biochem Biophys Res Commun. 1989;164:1–5. PMID:2480116. doi:10.1016/0006-291X(89)91813-5.
- Nijtmans LG, de Jong L, Sanz MA, Coates PJ, Berden JA, Back JW, Muijsers AO, van der Spek H, Grivell LA. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. Embo J. 2000;19:2444. PMID: 10835343. doi:10.1093/emboj/19.11.2444.
- Coates P, Nenutil R, McGregor A, Picksley SM, Crouch DH, Hall PA, Wright EG. Mammalian prohibitin proteins respond to mitochondrial stress and decrease during cellular senescence. Exp Cell Res. 2001;265:262. PMID:11302691. doi:10.1006/excr.2001.5166.
- Wang S, Fusaro G, Padmanabhan J, Chellappan S. Prohibitin co-localizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene. 2002;21:8388. PMID:12466959. doi:10.1038/sj.onc.1205944.
- Gamble S, Odontiadis M, Waxman J, Westbrook JA, Dunn MJ, Wait R, Lam EW-F, Bevan CL. Androgens target prohibitin to regulate proliferation of prostate cancer cells. Oncogene. 2004;23:2996. PMID: 14968116. doi:10.1038/sj.onc.1207444.
- Rajalingam K, Wunder C, Brinkmann V, Churin Y, Hekman M, Sievers C, Rapp UR, Rudel T. Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration. Nat Cell Biol. 2005;7:837–843. PMID: 16041367. doi:10.1038/ncb1283.
- Fusaro G, Dasgupta P, Rastogi S, Joshi B, Chellappan S. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J Biol Chem. 2003;278:47853–47861. PMID:14500729. doi:10.1074/jbc.M305171200.
- Coates PJ, Jamieson DJ, Smart K, Prescott AR, Biology PAHJC. The prohibitin family of mitochondrial proteins regulate replicative lifespan. Curr Biol. 1997;7:607–610. PMID: 9259555. doi:10.1016/s0960-9822(06)00261-2.
- Ikonen E, Fiedler K, Parton R, Simons K. Prohibitin, an antiproliferative protein, is localized to mitochondria. FEBS Letters. 1995;358:273–277. PMID: 7843414. doi:10.1016/0014-5793(94)01444-6.
- Terashima M, Kim KM, Adachi T, Nielsen PJ, Reth M, Köhler G, Lamers MC. The IgM antigen receptor of B lymphocytes is associated with prohibitin and a prohibitin-related protein. Embo J. 1994;13:3782–3792. PMID: 8070406.
- CS A, Lee JH, Reum Hwang A, Kim WT, Pai H-S. Prohibitin is involved in mitochondrial biogenesis in plants. Plant J. 2006;46:658–667. PMID:16640602. doi:10.1111/j.1365-313X.2006.02726.x.
- Van Aken O, Pecenková T, van de Cotte B, De Rycke R, Eeckhout D, Fromm H, De Jaeger G, Witters E, Beemster GTS, Inzé D, et al. Mitochondrial type-I prohibitins of Arabidopsis thaliana are required for supporting proficient meristem development. The Plant J. 2007;52:850–864. PMID: 17883375. doi:10.1111/j.1365-313X.2007.03276.x.
- Van Aken O, Whelan J, Van Breusegem F. Prohibitins: mitochondrial partners in development and stress response. Trends Plant Sci. 2010;15:275–282. PMID: 20226718. doi:10.1016/j.tplants.2010.02.002.
- Chen JC, Jiang CZ, Reid MS. Silencing a prohibitin alters plant development and senescence. Plant J. 2005;44:16–24. PMID:16167892. doi:10.1111/j.1365-313X.2005.02505.x.
- Wen T, Hochholdinger F, Sauer M, Bruce W, Schnable P. The roothairless1 gene of maize encodes a homolog of sec3, which is involved in polar exocytosis. Plant Physiol. 2005;138:1637–1643. PMID: 15980192. doi:10.1104/pp.105.062174.
- Christians MJ, Larsen PB. Mutational loss of the prohibitin AtPHB3 results in an extreme constitutive ethylene response phenotype coupled with partial loss of ethylene-inducible gene expression in Arabidopsis seedlings. J Exp Bot. 2007;58:2237–2248. PMID:17525078. doi:10.1093/jxb/erm086.
- Nadimpalli R, Yalpani N, Johal G, Simmons C. Prohibitins, stomatins, and plant disease response genes compose a protein superfamily that controls cell proliferation, ion channel regulation, and death. J Biol Chem. 2000;275:29579–29586. doi:10.1074/jbc.M002339200.
- Wang Y, Ries A, Wu K, Yang A, Crawford NM. The arabidopsis prohibitin gene phb3 functions in nitric oxide-mediated responses and in hydrogen peroxide-induced nitric oxide accumulation. Plant Cell. 2010;22:249–259. PMID:20068191. doi:10.1105/tpc.109.072066.
- Kong X, Tian H, Yu Q, Zhang F, Wang R, Gao S, Xu W, Liu J, Shani E, Fu C, et al. PHB3 maintains root stem cell niche identity through ROS-responsive AP2/ERF transcription factors in arabidopsis. Cell Rep. 2018;22:1350. PMID: 29386120. doi:10.1016/j.celrep.2017.12.105.
- Huang R, Shu S, Liu M, Wang C, Jiang B, Jiang J, Yang C, Zhang S. Nuclear prohibitin3 maintains genome integrity and cell proliferation in the root meristem through minichromosome maintenance. Plant Physiol. 2010;179:1669–1691. PMID: 30674698. doi:10.1104/pp.18.014632222.
- Seguel A, Jelenska J, Herrera-Vásquez A, Marr SK, Joyce MB, Gagesch KR, Shakoor N, Jiang S-C, Fonseca A, Wildermuth MC, et al. PROHIBITIN3 forms complexes with ISOCHORISMATE SYNTHASE1 to regulate stress-induced salicylic acid biosynthesis in arabidopsis. Plant Physiol. 2018;176:2515–2531. PMID: 29438088. doi:10.1104/pp.17.00941.
- Piechota J, Kolodziejczak M, Juszczak I, Sakamoto W, Janska H. Identification and characterization of high molecular weight complexes formed by matrix AAA proteases and prohibitins in mitochondria of Arabidopsis thaliana. J Biol Chem. 2010;23;285(17):12512–12521. PMID:20172857. doi:10.1074/jbc.M109.063644.
- Liu S, Yu F, Hu Q, Wang T, Yu L, Du S, Yu W, Li N. Development of in planta chemical cross-linking-based quantitative interactomics in arabidopsis. J Proteome Res. 2018;17:3195–3213. PMID: 30084631. doi:10.1021/acs.jproteome.8b00320.
- Wang W, Zhang X, Niittyla TJTPC. OPENER is a nuclear envelope and mitochondria localized protein required for cell cycle progression in arabidopsis. The Plant Cell. 2019;31:1446–1465. PMID: 31023726. doi:10.1105/tpc.19.00033.
- Choi D, Lee S, Hong S, Kim IH, Kang SMJO. Prohibitin interacts with RNF2 and regulates E2F1 function via dual pathways. Oncogene. 2008;27:1716–1725. PMID: 17873902. doi:10.1038/sj.onc.1210806.
- Diego JGD, Rodríguez FD, Lorenzo JLR, Physiology ECJJOP. The prohibitin genes in Arabidopsis thaliana: Expression in seeds, hormonal regulation and possible role in cell cycle control during seed germination. J Plant Physiol. 2007;164:371–373. PMID: 16876910. doi:10.1016/j.jplph.2006.05.002.
- Van Aken O, Ford E, Lister R, Huang S, Millar AHJPJ. Retrograde signalling caused by heritable mitochondrial dysfunction is partially mediated by ANAC017 and improves plant performance. Plant J. 2016;88:542–558. PMID: 27425258. doi:10.1111/tpj.13276.
- Zhang S, Wu J, Yuan D, Zhang D, Huang Z, Xiao L, Yang C. Perturbation of auxin homeostasis caused by mitochondrial ftsh4 gene-mediated peroxidase accumulation regulates arabidopsis architecture. Mol Plant. 2014;7:856–873. PMID: 24482432. doi:10.1093/mp/ssu006.
- Zhang S, Li C, Wang R, Chen Y, Shu S, Huang R, Zhang D, Li J, Xiao S, Yao N, et al. The arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY-dependent salicylic acid accumulation and signaling. Plant Physiol. 2017;173:2294–2307. PMID: 28250067. doi:10.1104/pp.16.00008.
- Wei Y, Chiang W-C, Sumpter R, Mishra P, Levine BJC. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell. 2017;168:224. doi:10.1016/j.cell.2016.11.042.
- Junior TCT, de Godoy LMF, de Souza GA, Bonatto D, Otake AH, de Freitas Saito R, Rosa JC, Greene LJ, Chammas R. Accumulation of prohibitin is a common cellular response to different stressing stimuli and protects melanoma cells from ER stress and chemotherapy-induced cell death. Oncotarget. 2017;8:43114–43129. PMID: 28562344. doi:10.18632/oncotarget.17810.
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GTS, Gruissem W, Van de Peer Y, Inzé D, De Veylder L. Genome-wide identification of potential plant E2F target genes. Plant Physiol. 2005;139:316–328. PMID: 16126853. doi:10.1104/pp.105.066290.
