ABSTRACT
There is a scarcity of research reports on the effect of ultraviolet (UV)-B radiation on genome-wide transcriptional regulation in the multicellular green microalga including Volvox carteri (V. carteri). This microalga possesses only two cell types including mortal and motile somatic cells, as well as immortal and immotile reproductive cells. Therefore, the present study evaluated the effect of low-dose UV-B radiation on the cell-type-specific gene expression pattern of reproductive and somatic cells in an asexual life cycle of V. carteri using RNA sequence method. To this end, the separated reproductive and somatic cells were treated for 1 hour at an intensity of 0.056 mW/cm−2 UV-B radiation. Then, a transcriptome analysis was conducted between the UV-B and white light treated groups in either of the cell types. Based on differential gene expression analyses, no differentially expressed genes were found in reproductive cells under the treatment as compared to the control group. This type of cell maintained its steady state. However, treating the somatic cells with UV-B radiation led to at least 126 differentially expressed genes compared to the untreated control group. In addition, the results of a direct comparison demonstrated a restricted and wide response to UV-B radiation in somatic cells as compared to reproductive cells. Based on the results, UV-B radiation could be involved in cell-type-specific regulation of biological pathways.
Introduction
The sunlight that reaches the surface of the earth contains ultraviolet (UV)-B radiation (280–315 nm), UV-A radiation (315–400 nm), photosynthetically active radiation (PAR, 400–700 nm), and infrared radiation (>700 nm) Citation1. A trivial increase in UV-B radiation on the earth surface may lead to unbalance natural ecosystem. Therefore, UV-B, among the various types of UV radiations, is of great consideration.Citation2
UV-B exerts complex effects on plants. Such effects are studied to induce defense mechanisms or photomorphogenic responses in a range of higher plants such as Arabidopsis thaliana, Brassica rapa, Glycine max, Vitis vinifera, as well as in lower plants like Umbilicaria aprina, Bryum argenteum, Kappaphycus alvarezii, C. reinhardtii, Micromonas commode and Chlorella sp.Citation3–Citation11 According to these studies, UV-B radiation generally affects signaling, the regulation of gene expression, and biological pathways in plants. However, other model plant species should be investigated to better understand these processes. Despite many studies on green microalgae, so far, they have all investigated the effect of UV-B radiation only on unicellular species. Therefore, there is a significant knowledge gap respecting understanding the acquisition and regulation of UV-B tolerance in multicellular green microalgae at the genome-wide level. The multicellular green alga Volvox carteri is a model organism for developmental, physiological and evolutionary research.Citation12 Thus, by delving into responsive mechanisms of UV-B tolerance help to strengthen the Volvox genome annotation as a model organism and present an overview of the gene expression pattern in multicellular green algae.
The monophyletic group of volvocine contains multiple algae species ranging from the unicellular type of Chlamydomonas reinhardtii to the multicellular genus Volvox. The unicellular green flagellate alga C. reinhardtii is a well-studied model organism which is commonly used as an outgroup to the multicellular speciesCitation13,Citation14 and is considered as a reference point for evaluating the differences and similarities underlying unicellular and multicellular organizations.Citation15
Volvox carteri (V. carteri) is a haploid, photoautotrophic freshwater green alga with large colonies (up to 1.5 mm) that encompass only two cell types.Citation16,Citation17 The large reproductive cells (gonidia) are immotile, potentially immortal and functionally specialized for growth and the reproduction of new colonies while the small somatic cells with terminal differentiation are specialized for support functions (e.g., phototactic motility). These cells are programmed to senesce and die when they are only a few days old.Citation16–Citation18
Many types of photoreceptors are identified in V. carteri, which have different expressions in reproductive and somatic cells. This multicellular green alga seems to have a photoreceptor which can play a role in UV-B response.Citation19 According to previous studies, reproductive and somatic cells demonstrate a differentially expressed gene (DEG) pattern from various functional classes.Citation20,Citation21
Transcriptome profiling is utilized as one of the most frequent approaches for investigating the changes in gene expression at the molecular level. Recently, studies on transcriptome gene expression have investigated the cell-type-specific gene expression.Citation12,Citation22 However, these studies were performed under the conditions of photosynthetically active radiation (PAR, 400–700 nm).
The current study sought to examine the effect of low-dose UV-B radiation on the cell-type-specific gene expression pattern of reproductive and somatic cells in an asexual life cycle of V. carteri employing a comprehensive transcriptome analysis by an RNA-Seq method.
Results
RNA isolation, RNA-seq libraries and mapping
A total of ~276 and ~269 million raw reads were generated for reproductive and somatic cells, respectively. After processing the raw reads (i.e., short and low-quality reads) for adapter contamination, a total of 272 and 264 million clean reads were screened for reproductive and somatic cells, respectively. The clean reads were then mapped onto the V. carteri genome assembly using TopHat software. As a result, almost 91.9–94.9% of the clean reads were mapped on the reference genome. Detailed information related to the raw and cleaned RNA-Seq reads, as well as the mapping percentage of the reads on the genome, are presented in .
Table 1. General information regarding RNA-Seq data for both reproductive and somatic cells of V. carteri under UV-B treated or control conditions.
Comparative analysis of differential gene expression in control and UV-B treated samples
To detect the possible gene expression modifications between the UV-B and white light treated groups in either of the cell types, the significant differential expression level of gene features including gene, isoform, transcription start site (TSS) and coding sequences (CDS) was compared using normalized RPKM values by Cufflinks software. Furthermore, a total of 24089 genes, 50155 isoforms, 35036 TSS and 15206 CDS were assessed for reproductive cell type. Surprisingly, no differentially expressed gene features were observed (Table S1). Moreover, 23015 genes, 46879 isoforms, 32940 TSS and 15207 CDS groups were assayed for somatic cell type. Based on the results, 126 genes, 72 isoforms, 89 TSS and 44 CDS were differentially expressed under UV-B treatment. ) and () displays the number of differentially expressed genes (Table S2 (a) and (b)). As shown, 102 (out of 126) genes, 60 (out of 72) isoforms, 74 (out of 89) TSS and 40 (out of 44) CDS are overexpressed in the UV-B-treated group compared to the control group, suggesting a great stimulating effect of UV-B on the function of some genes. In other words, based on the obtained results, about 81% and 19% of genes from 126 differentially expressed genes are up- and down-regulated, respectively. Although all the gene features can be discussed comprehensively, only differentially expressed genes are analyzed to avoid reader confusion. Additionally, a volcano plot of the distribution of significantly regulated genes is shown in . Furthermore, a heatmap () is provided to illustrate the differential gene expression variations under treatment among different replicates. As depicted, all significantly expressed loci with no annotations are considered novel and marked with novel prefix in the heatmap.
Figure 1. (a) The number of differentially up- and down-regulated gene features between treatment (UV-B) and control (WL) groups in somatic cells. Note. N is the total number of gene features with and New represents the number of unannotated gene features; (b) Differentially expressed genes (i.e., 0–1, 1–2, & >2 log2 fold change) in somatic cells.
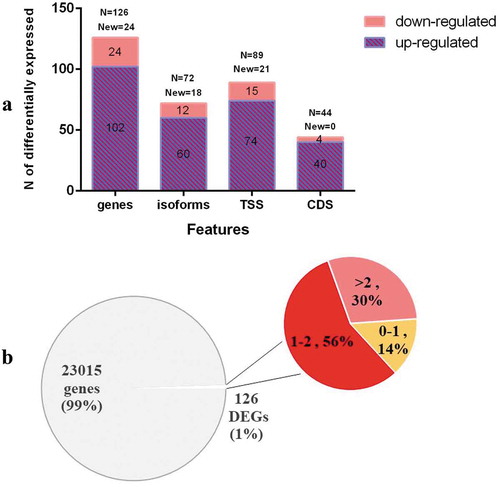
Figure 2. The volcano plot of the transcriptome in somatic cells under UV-B and WL treatment.
Note. The genes above the horizontal line are significantly based on a threshold (FDR<0.05) and the two-fold change threshold (|log2FoldChange|>1), respectively.
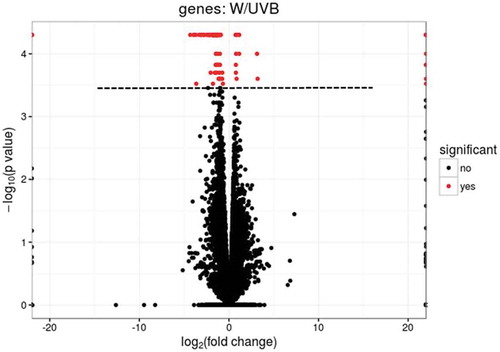
Figure 3. Heatmap (log RPKM+1) showing the expression profiles of differentially expressed genes between UV-B treated (UVB) and white light control (w) groups in somatic cells.
Note. Due to non-stranded library to generate the data, some loci included two known genes that are marked with integer number postfixes on the graph. These loci are as below:
Investigating the expression of UV-B photoreceptor and target genes of UV-B response pathway
The analysis of transcript level in reproductive and somatic cells showed that the expression level of VcUVR8‑V1 gene (UV-B photoreceptor) failed to significantly differ under the low intensity of UV-B radiation (Figure S1 and S2 and Table S3). In addition, the expression of the candidate genes of the UV-B response pathway represented no significant difference between the treated and control groups following investigating these genes in reproductive cells (Figure S3). ABC transporter family protein was observed in somatic cells under UV-B treatment as well (Figure S4 and Table S4).
Gene ontology (GO) and kyoto encyclopedia of gene and genome (KEGG) pathway analysis in somatic cells
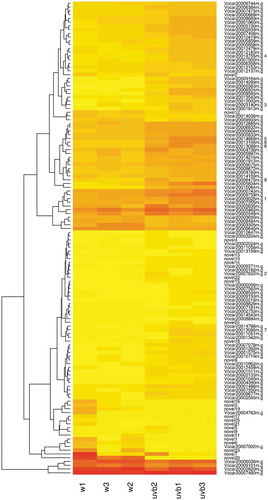
Algal Functional Annotation web-tool was used for functional annotation and classification of 126 differentially expressed genes (DEGs) in somatic cells of V. carteri. Based on the analysis, 68 out of 126 DEGs in somatic cells of V. carteri were obtained as homologous genes in C. reinhardtii. These 68 homologous genes were subjected to enrichment analysis of GO and biological pathways using Algal Functional Annotation Tool. GO terms were analyzed based on the Joint Genome Institute (JGI) annotation to obtain GO classifications according to biological process, molecular function and cellular component. Pathway enrichment was analyzed by the KEGG database. Based on JGI annotation, the results of GO annotation of 68 homologous genes revealed that, in biological process category, the assignments were prominently enriched in the terms of phytochromobilin biosynthetic and metabolic processes, the regulation of lyase activity, alanyl-tRNA aminoacylation, gluconeogenesis, rRNA modification, isoprenoid biosynthetic and metabolic processes, as well as hexose and monosaccharide biosynthetic processes. Their molecular function category was primarily related to “oxidoreductase activity, acting on the CH-CH group of donors, iron-sulfur protein as an acceptor”, 2-amino-5-formylamino-6-(5-phosphoribosylamino)pyrimidin-4(3H)-one formate-lyase activity, cobalt ion binding, phosphoenolpyruvate carboxykinase (ATP) activity, alanine-tRNA ligase activity, epoxide hydrolase activity, tocopherol O-methyltransferase activity, “all-trans-retinol 13,14-reductase activity”, ether hydrolase activity, phosphoenolpyruvate carboxykinase activity, “oxidoreductase activity, acting on the CH-CH group of donors” and “hydrolase activity, acting on ether bonds”. In the present study, there are no significant GO terms in the cellular component category. The results of GO enrichment analysis are shown in (Table S7).
Figure 4. Functional analysis of DEGs between the UV-B treatment and control groups in the somatic cells based on gene ontology. GO category was presented for two major functional categories, namely biological process and molecular function.
KEGG pathways were analyzed to identify the active biological pathways in the somatic cells of V. carteri And 68 homologous genes were assigned to 22 KEGG pathways. Five pathways were significantly (P < .05) affected including peroxisome proliferator-activated receptors (PPAR) signaling pathway, aminoacyl-tRNA biosynthesis, porphyrin and chlorophyll metabolism, vitamin B6 metabolism and alpha-linolenic acid metabolism (Table S8).
PPAR signaling pathway
(Figure S5): These genes encode for acyl-CoA oxidase (EC: 1.3.3.6) and solute carrier family 25 (mitochondrial uncoupling protein), member 7 (UCP1). The acyl-CoA oxidase (Acox) and UCP1 are fatty acid oxidation-associated enzymes. Therefore, the breakdown of fatty acids is a result of UV-B treatment. In this pathway, two genes were up-regulated in response to UV-B treatment.
Aminoacyl-tRNA biosynthesis
(Figure S6): The alanyl-tRNA synthetase (alaS) is a ligase which catalyzes the attachment of alanine group to the hydroxyl of tRNA(Ala) and forms carbon-oxygen bonds in aminoacyl-tRNA. The gene coding alanyl-tRNA synthetase (EC: 6.1.1.7) was up-regulated in response to UV-B treatment.
Porphyrin and chlorophyll metabolism
(Figure S7): These genes encode pheophorbide a oxygenase (EC: 1.14.15.17) and phycocyanobilin:ferredoxin oxidoreductase (EC: 1.3.7.5). Pheophorbide a oxygenase is involved in the pathway chlorophyll degradation. Phycocyanobilin, namely, ferredoxin oxidoreductase catalyzes the biosynthesis of the phycobiliprotein. Two genes were up-regulated in response to UV-B treatment in this pathway.
Vitamin B6 metabolism
(Figure S8): Pyridoxal 5ʹ-phosphate synthase pdxS subunit (EC: 4.3.3.6) was up-regulated in this vitamin metabolism pathway in response to UV-B treatment.
Alpha-linolenic acid metabolism
(Figure S9): The acyl-CoA oxidase (EC: 1.3.3.6) was up-regulated in this metabolism pathway.
Discussion
Through using the comprehensive transcriptome analysis through RNA-Seq technique, the direct effects of low-dose UV-B radiation (0.056 mW.cm−2) on the general changes of gene expression (transcriptome profile) were investigated in separated cell-types of V. carteri in the vegetative life cycle.
Investigating the expression of UV-B photoreceptor and target genes of the UV-B response pathway
VcUVR8-V1 gene (UV-B photoreceptor) showed no significant differential expression either in reproductive or somatic cells in comparison with their corresponding control groups. So far, many studies have demonstrated that UV-B radiation changes no protein abundance of UVR8 while it induces the monomerization and nuclear accumulation of UVR8.Citation6,Citation23 Conversely, the expression of the ABC transporter family among the target genes of the UV-B response pathway was significant in somatic cells. Recent findings indicated that the members of this family contribute to homeostasis processes after UV-B radiation.Citation24
The response pattern of cell types to UV-B treatment
Unlike somatic cells, no effect of one h UV-B was observed on the regulation of gene expression in reproductive cells. Probably, reproductive cells possess differentially UV absorptive properties, which may serve to decrease the effects of UV-B on such cells.Citation25 The post-transcriptional control by regulators including small non-coding (silencing) RNAs may be highlighted for the lack of differential expression in reproductive cells.Citation26 Furthermore, this could be due to their function in the production of the next generation. Hence, these cells should be more conserved in facing environmental conditions including UV-B radiation. Based on the obtained results, unlike reproductive cells, somatic cells were readily programmed to respond to UV-B light exposure during their evolution procedure.
Candidate biological pathways and enzymes in somatic cells during UV-B treatment
Limitations were implemented on the genes annotation as the result of the annotation of available green microalgal genomes which was not complete. Therefore, due to the weaknesses of software and a poor annotation of V. carteri, information of its close unicellular relative C. reinhardtii, represented in the Algal Functional Annotation Tool,Citation27 was applied in the present study.
In the volvocine algae, C. reinhardtii is typically utilized as an outgroup for the multicellular speciesCitation14,Citation28 and is regarded as a reference point for investigating the differences and similarities underlying unicellular and multicellular organizations.Citation15 In the current study, 68 out of 126 differentially expressed genes in somatic cells of V. carteri were obtained as homologous genes in C. reinhardtii. Then, these homologous genes were assigned to 22 KEGG pathways. Based on the analysis, five metabolic pathways were significantly affected by UV-B treatment (P-value<0.05) and the enriched genes in these biological pathways were up-regulated accordingly.
Peroxisome proliferator-activated receptor signaling pathway and alpha-linolenic acid metabolism
The Acox and UCP1 genes were enriched in these biological pathways, which were related to the oxidation of various fatty acids. PPARs act as lipid metabolism key transcriptional regulators, antioxidant defense and mitochondrial biogenesis.Citation29 Furthermore, the expression of the genes networks are controlled by PPARs; they can encode the proteins which are available in the whole process of lipid metabolism.Citation30 In addition, the genes which play the role in regulation of lipid homeostasis were controlled by these transcription factors.Citation31 Alpha-linolenic acid (ALA) functions as the predecessor of oxidized fatty acid metabolites called oxylipins in plant or algae oilsCitation32 As a defense process against the pathogens, oxylipins in the plants which are derived from ALA are utilized.Citation33,Citation34 In addition, the expression of an acyl‐CoA oxidase gene of β‐oxidation was found to be increased by the UV light; it suggests that for a secondary metabolism, the pathway can be used in preparation of acetyl‐CoA substrate.Citation35 Therefore, reacting to membrane lipid peroxidation and regarding specific cellular metabolic conditions and based on repair functions, the cells might cause cell death or they may lead to successful cell survival.Citation36 Lipids are involved in the mitigation processes; this is its feedback to imposed stress on the plant cellsCitation37,Citation38 while they are also engaged in the start of defense reactions as intermediates of signals.Citation39,Citation40 Via various dynamics of lipids, membrane integrity and the functions of membrane‐bound protein, remodeling of the membrane lipid plays a significant role in the conserving membrane features; this remodeling is a response to stresses namely dehydration, freezing and nutrition depletion.Citation39,Citation41,Citation42 Energy supply and energy demand of cells are influenced by UV-B radiation. For regulation of thermogenesis and expenditure of energy, UCP1 acts as mitochondrial carrier and changes the expression of chloroplast genes which causes expression of a large subset of stress-responsive genes providing the situation to clarify the stress tolerance regarding those biotic or abiotic ones.Citation35
Aminoacyl-tRNA biosynthesis
alaS gene was enriched in this biological pathway. Protein alterations can be promoted by free radicals which have their source in UV-B radiation.Citation43 A wide variety of conditions and various cellular compartments are required to be sure of an exact and efficient synthesis process of protein by Aminoacyl-tRNA synthetases.Citation44 RNA ligases play an important role both in processing the RNA transcripts with different origins and in RNA repair pathways.Citation45,Citation46 Some responses to stress were modulated by the translation system,Citation47 but the problem in understanding the effects of UV-B radiation on any of the components of the translation system is due to lack of information.
Porphyrin and chlorophyll metabolism
In the photosynthetic light-harvesting structures of cyanobacteria which are called phycobilisomes, the light-harvesting pigment is phycocyanobilin.Citation48 Photosynthesis is possible even under dim light by the permission of Phycobilin-containing phycobilisomes. On the other hand, when more intense light is available than the light which can be efficiently used in generating the reduced carbon, cells can be made vulnerable by phycobilisomes; this is the difference with moderate efficient light that makes the reduced carbon produced.Citation49,Citation50 Pheophorbide a oxygenase (PAO) gene is enriched in chlorophyll metabolism. Additionally, UV-B radiation produces free radicals participating in the degradation of chlorophyll and leads to the concomitant destruction of the photosynthetic machinery.Citation51 Therefore, photosynthetic reaction centers and antenna complexes are distinguished as primordial UV-protectors of the cell.Citation52 Besides, the main point in the control of the regulation of chlorophyll degradation seems to be PAO which its function is to catalyze the oxygenic ring opening of pheophorbide.Citation53,Citation54 In addition, the consonant breakdown of chlorophyll by PAO enzyme maintains the viability of cells within abiotic stresses including oxidative stress by UV-B radiation.Citation55
Vitamin B6 metabolism
pdxS gene was enriched in vitamin B6 metabolism. This vitamin is soluble in water and serves as a cofactor for a wide group of essential enzymes which are chiefly connected to synthesis of amino acid.Citation56,Citation57 Also, antioxidant activities are exhibited by vitamin B6 and it plays a significant role in cell protection against oxidative stress.Citation58 Researches on Arabidopsis show that vitamin B6 is linked to responses to stress. Indeed, it is important for acclimation to osmotic and oxidative stressCitation59 and functions efficiently in photoprotection.Citation60
The different cell types have various structures and are designed to perform specific functions. Therefore, they differ in terms of energy balance.Citation20,Citation22 Apparently, the effects of UV-B radiation can be prohibited and reduced in the reproductive cells, and therefore, healthy cells can produce the next generation. In the present study, the reproductive cell-type maintained its steady state. The obtained results indicated a restricted but widespread response of somatic cells to UV-B radiation since several biological pathways interact in somatic cells. Additionally, somatic cells seem to express more specialized functions and programs compared to reproductive cells.
Conclusion
In general, the findings of the current study confirmed the influence of low-dose UV-B radiation on the cell-type-specific gene expression. The cell-type-specific genes were identified in response to the applied radiation. The reduced effects of UV-B on reproductive cells could be due to their function in creating the next generation. Accordingly, they should be more conserved when encountering environmental conditions including the UV-B radiation. In addition, a total of 126 differentially expressed genes were identified among the treatment and control groups in somatic cells. The results further indicated that, under low-dose UV-B radiation, somatic cells overexpress some genes in several metabolic pathways, especially peroxisome proliferator-activated receptor signaling pathway, in order to preserve cell homeostasis and increase cellular performance. Based on the results, somatic cells expressed more specialized programming and functionality. Finally, the genes of reproductive cells were unexpressed in response to low-dose UV-B and thus should be subject to further investigation at higher doses of UV-B radiation.
Materials and methods
Strains and culture conditions
The wild-type V. carteri f. nagariensis female strain Eve10Citation61 is the descendant of HK10,Citation62 which originates from Japan. In the present study, cultures were grown synchronously under vegetative conditions using Standard Volvox Medium (SVM)Citation63 at 28ºC under a 16:8-h light-to-dark cycleCitation64 with average illumination of ~100 μmol photons m−2 s−1 PAR in the glass Erlenmeyer flask that possessed caps for madding the age exchange.
Separation of cell types
In a short time before at the start of the action of cleaving which happens in regenerative cells with the use of filtering tools, Volvox spheroids got gathered; these spheroids’ volume was determined to be 10 l while Volvox spheroids were going through the natural process of growth at the same time; it should be noted that SVM was utilized to bring the concentration of organisms to approximately 1000 spheroids/ml. Moreover, in a special process which involves the pestle going up and down, with pestle which is fit completely (B. Braun, Melsungen, from Germany), the spheroids were separated into parts in a 50 ml Dounce homogenizer; it was done to have reproductive cells. Then, 500 rpm in 5 min was chosen as arrangements of centrifuging to keep the reproductive cells apart from the majority of the group; the group included residual somatic cells and extracellular matrix fragments (Sigma-Aldrich, St. Louis, MO). As the next step, filtration was done on nylon screen with a thickness of 10 μm and the point is that it was not possible for the reproductive cells to make their ways through the screen. With the use of SVM as much as 100 ml, the screen was cleaned from the reproductive cells; washing the screen was done 3 times thus preparing gonidial RNA process was done. With the use of a completely fitted pestle which 7 times was taken up and down, the breaking process of spheroids in the 50 ml Dounce homogenizer was done so that somatic cells be achieved. As a result, the SVM was used to dilute the achieved cell suspension to the two-fold volume one. Unit gravity was then applied for 20 min to settle the reproductive cells and larger fragments of spheroids which included the reproductive cells. In contrast, somatic cell sheets were then taken and inserted to the somatic RNA preparation while there were no reproductive cells floating on the surface.Citation20
UV-B treatment
Cell types were isolated before initiating cleavage division and then they were illuminated with UV-B light (0.056 mW/cm2 UV-B [Philips TL40W/12RS], measured with a PCE-UV34 UVA/UVB radiation meter, PCE Instruments) supplemented with 0.8 μmol m−2 s−1 white light (measured with the LI-250 Light Meter, LI-COR Biosciences) for one h. Finally, the control cells were incubated under UV exclusion as well.
Total RNA extractions for next-generation sequencing
Three biological replicates per treatment and cell types (totally 12 samples) were used for RNA extraction using TRIzol Reagent (Sigma-Aldrich, St. Louis, MO) according to the manufacturer’s instructions. The isolated RNA was checked for purity and quantity using 1% agarose gel electrophoresis and Ultrospec 2100 pro UV/Visible spectrophotometer (GE Healthcare, Uppsala, Sweden). Furthermore, the RNA concentration was adjusted to 100 ng/μL and the quality was re-checked by an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). All 12 RNA samples passed both quality thresholds, namely, the 260/280 odds ratios were between 1.8 and 2.2 and the RNA integrity number was above 8.
Library preparation and massively parallel sequencing
The total RNA of 12 samples was submitted to the GATC Biotech (Konstanz, Germany) for Illumina-based polyA library preparation and sequencing. Briefly, one μg of the total RNA of each sample was exposed to poly (A)+ mRNA enrichment. The poly-A enriched mRNAs were fragmented and then, cDNA was synthesized by random primer, followed by ligating to Illumina sequencing adapters. Eventually, the sequencing runs were performed on an Illumina HiSeq-2500 in order to generate 51 base single-end reads. The employed RNA-Seq data were recorded in the Sequence Read Archive in the National Center for Biotechnology Information (NCBI, https://blast.ncbi.nlm.nih.gov) with a GSE124346 access number.
Transcriptome analysis
The quality control and preprocessing of the sequenced reads were evaluated using FastQCCitation65 and Trimmomatics (version 32)Citation66 instruments, respectively. Briefly, adapter contamination was checked and then, unknown or low-quality bases (with a quality score <Q25) were trimmed as well. Furthermore, reads with an average quality score below 25 or with a length smaller than 25 were filtered out. All analyses were performed using quality-checked clean data. Then, the reads of 12 samples were aligned onto V. carteri genome assembly (available in Phytozome; https://phytozome.jgi.doe.gov) (version 2)Citation67 using TopHat2 softwareCitation68 utilizing – N 1 – coverage-search – read-realign-edit-dist 0 g 1 parameters. Following alignment, the mapped reads were visualized by the Integrative Genomics Viewer software (version 2.1)Citation69 in order to check and evaluate the coverage of the aligned data. Then, the transcripts per sample were assembled employing Cufflinks software (version 2.0.1)Citation70 with parameters of – p 2 – max-frag-multihits 1. Next, these transcripts were merged using cuffmerge, which is an accompanying program of the Cufflinks package. Moreover, Cuffdiff software (version 2) was used to test statistically significant differences in transcript abundance between the control and treatment groups of both cell types, separately. The criteria for discovering the significant differentially expressed genes (DEGs) included a minimum alignment count of 10 reads, absolute fold-change ≥ 2, and q-value (false discovery rate-adjusted P-value) <0.05. Finally, RStudio software (version 3.3.0) was applied to run R scripts to accomplish hierarchical cluster analysis of DEGs expression and to create heatmap using appropriate packages.
Investigating the expression of UV-B photoreceptor and target genes of the UV-B response pathway
The sequence of VcUVR8‑V1 gene in V. carteri was used to investigate UV-B photoreceptor.Citation19 In addition, a set of candidate genes was selected from different gene databases such as NCBI, Phytozome (phytozome.jgi.doe.gov) and Ensembl (plants.ensembl.org) based on the available observations from Arabidopsis.Citation71,Citation72 Eventually, the above-mentioned genes were chosen to monitor the modification of different cell-type specific light signaling pathways in response to UV-B light in order to investigate their existence under UV-B treatment.
Annotation
The reference genome annotation files of V. carteri f. nagariensis were obtained from the Phytozome V12 platform.Citation73 Due to the weaknesses of the software and a poor annotation of V. carteri, the Algal Functional Annotation ToolCitation27 was used for Gene Ontology (GO) and biological pathway analysis. This software currently covers no alga V. carteri, therefore, the information of its close unicellular relative C. reinhardtii was employed instead. Following differential gene expression analysis using cufflinks software, the assembled transcripts were annotated using BLAST alignment in the NCBI non-redundant protein databaseCitation74 with an – E-value ≤ 1e – 8. Indeed, the accession numbers with the prefix XP_(protein) were obtained for V. carteri from NCBI data using the BLASTX tool. Further, the homologous genes of V. carteri in C. reinhardtii alga were identified by using the BLASTP instrument. The accession numbers with the prefix XM_(mRNA), XP_(protein), and in some cases, ID Phytozome of C. reinhardtii were obtained as well (Table S5). Then, the obtained accession numbers with the prefix XM_(mRNA) and ID Phytozome were used in the web application software of Algal Functional Annotation Tool (Table S6). Based on Joint Genome Institute annotation, Gene Ontology (GO) terms were analyzed to obtain GO classifications according to biological process, molecular function and cellular component, respectively. Furthermore, pathway enrichment was analyzed using the Kyoto Encyclopedia of Genes and Genomes database. Then, the enrichment results were sorted by hypergeometric P-value in the Algal Functional Annotation Tool.Citation27 The threshold of P-value was reported <0.05 for the selection of the overrepresented enrichment results.
Abbreviations
| RNA-Seq | = | ribonucleic acid sequencing |
| MSI | = | Mass spectrometry imaging |
| PAR | = | photosynthetically active radiation |
| ROS | = | reactive oxygen species |
| SVM | = | Standard Volvox Medium |
| UV-B | = | Ultraviolet B |
| DEGs | = | differentially expressed genes |
| ECM | = | extracellular matrix |
| NCBI | = | National Center for Biotechnology Information |
| SRA | = | Sequence Read Archive |
| GO | = | gene ontology (database) |
| KEGG | = | Kyoto Encyclopedia of Genes and Genomes |
| ACO | = | acyl-CoA oxidase |
| UCPs | = | mitochondrial uncoupling proteins |
| PPARα | = | peroxisomal proliferator-activated receptor α |
| alaS | = | Alanyl-tRNA biosynthesis |
| PAO | = | pheophorbide a oxygenase |
| PcyA | = | Phycocyanobilin:ferredoxin oxidoreductase |
| pdxS | = | pyridoxal 5’-phosphate synthase |
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Competing interests
The authors declare that they have no competing interests.
Supplemental Material
Download Zip (5.2 MB)Acknowledgments
We would like to thank all the publicly available databases and the scientists behind them. Special thanks go to Girish Beedessee for manuscript revision and helpful suggestions.
Supplemental Material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Kollias N, Ruvolo E, Sayre RM. The value of the ratio of UVA to UVB in sunlight. Photochem Photobiol. 2011;87(6):1–11. doi:10.1111/j.1751-1097.2011.00980.x.
- Sztatelman O, Grzyb J, Gabryś H, Banaś AK. The effect of UV-B on Arabidopsis leaves depends on light conditions after treatment. BMC Plant Biol. 2015;15(1):281. doi:10.1186/s12870-015-0667-2.
- Cuvelier ML, Guo J, Ortiz AC, Van Baren MJ, Tariq MA, Partensky F, Worden AZ Responses of the picoprasinophyte Micromonas commoda to light and ultraviolet stress. PLoS One. 2017;12(3):e0172135. Cockshutt AM, ed. doi:10.1371/journal.pone.0172135.
- Loyola R, Herrera D, Mas A, Wong DCJ, Höll J, Cavallini E, Amato A, Azuma A, Ziegler T, Aquea F, et al. The photomorphogenic factors UV-B RECEPTOR 1, ELONGATED HYPOCOTYL 5, and HY5 HOMOLOGUE are part of the UV-B signalling pathway in grapevine and mediate flavonol accumulation in response to the environment. J Exp Bot. 2016;67(18):5429–5445. doi:10.1093/jxb/erw307.
- Poong SW, Lim PE, Phang SM, Wong C-Y, Pai T-W, Chen C-M, Yang C-H, Liu -C-C. Transcriptome sequencing of an Antarctic microalga, Chlorella sp. (Trebouxiophyceae, Chlorophyta) subjected to short-term ultraviolet radiation stress. J Appl Phycol. 2018;30(1):87–99. doi:10.1007/s10811-017-1124-4.
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, et al. Perception of UV-B by the arabidopsis UVR8 protein. Science. 2011;332(6025):103–106. doi:10.1126/science.1200660.
- Schmidt ÉC, Nunes BG, Maraschin M, Bouzon ZL. Effect of ultraviolet-B radiation on growth, photosynthetic pigments, and cell biology of Kappaphycus alvarezii (Rhodophyta, Gigartinales) macroalgae brown strain. Photosynthetica. 2010;48(2):161–172. doi:10.1007/s11099-010-0022-7.
- Singh J, Singh R. Adverse effects of UV-B radiation on plants growing at schirmacher oasis, East Antarctica. Toxicol Int. 2014;21(1):102. doi:10.4103/0971-6580.128815.
- Tilbrook K, Dubois M, Crocco CD, Yin R, Chappuis R, Allorent G, Schmid-Siegert E, Goldschmidt-Clermont M, Ulm R. UV-B Perception and Acclimation in Chlamydomonas reinhardtii. Plant Cell. 2016;28(4):966–983. doi:10.1105/tpc.15.00287.
- Wang J, Wang Y, Chen BW, Kawabata S, Li Y. Comparative transcriptome analysis revealed distinct gene set expression associated with anthocyanin biosynthesis in response to short-wavelength light in turnip. Acta Physiol Plant. 2016;38(6):134. doi:10.1007/s11738-016-2145-3.
- Yoon MY, Kim MY, Shim S, Kim KD, Ha J, Shin JH, Kang S, Lee S-H. Transcriptomic profiling of soybean in response to high-intensity UV-B irradiation reveals stress defense signaling. Front Plant Sci. 2016;7:(December):1–6. doi:10.3389/fpls.2016.01917.
- Klein B, Wibberg D, Hallmann A. Whole transcriptome RNA-Seq analysis reveals extensive cell type-specific compartmentalization in Volvox carteri. BMC Biol. 2017;15(1):111. doi:10.1186/s12915-017-0450-y.
- Coleman AW. Phylogenetic analysis of “Volvocacae” for comparative genetic studies. Proc Natl Acad Sci U S A. 1999;96(24):13892–13897. doi:10.1073/pnas.96.24.13892.
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318(5848):245–250. doi:10.1126/science.1143609.
- Nishii I, Miller SM. Volvox: simple steps to developmental complexity? Curr Opin Plant Biol. 2010;13(6):646–653. doi:10.1016/j.pbi.2010.10.005.
- Hallmann A. Evolution of reproductive development in the volvocine algae. Sex Plant Reprod. 2011;24(2):97–112. doi:10.1007/s00497-010-0158-4.
- Kirk DL. A twelve-step program for evolving multicellularity and a division of labor. BioEssays. 2005;27(3):299–310. doi:10.1002/bies.20197.
- Michod RE, Viossat Y, Solari CA, Hurand M, Nedelcu AM. Life-history evolution and the origin of multicellularity. J Theor Biol. 2006;239(2):257–272. doi:10.1016/j.jtbi.2005.08.043.
- Razeghi J, Kianianmomeni A. UV-B response is modulated by cell-type specific signaling pathway in multicellular green algae Volvox carteri. Plant Growth Regul. 2019;87(2):303–315. doi:10.1007/s10725-018-0472-7.
- Nematollahi G, Kianianmomeni A, Hallmann A. Quantitative analysis of cell-type specific gene expression in the green alga Volvox carteri. BMC Genomics. 2006;7(1):321. doi:10.1186/1471-2164-7-321.
- Tam LW, Kirk DL. Identification of cell-type-specific genes of Volvox carteri and characterization of their expression during the asexual life cycle. Dev Biol. 1991;145(1):51–66. doi:10.1016/0012-1606(91)90212-L.
- Matt GY, Umen JG. Cell-type transcriptomes of the multicellular green alga volvox carteri yield insights into the evolutionary origins of germ and somatic differentiation programs. G3: Genes|Genomes|Genetics. 2017;8(2):531–550. doi:10.1534/g3.117.300253.
- Kaiserli E, Jenkins GI. UV-B promotes rapid nuclear translocation of the arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell Online. 2007;19(8):2662–2673. doi:10.1105/tpc.107.053330.
- Hwang JU, Song WY, Hong D, Ko D, Yamaoka Y, Jang S, Yim S, Lee E, Khare D, Kim K, et al. Plant ABC transporters enable many unique aspects of a terrestrial plant’s lifestyle. Mol Plant. 2016;9(3):338–355. doi:10.1016/j.molp.2016.02.003.
- Jacquemoud D, Pohnert G. Extraction and analysis of oxylipins from macroalgae illustrated on the example Gracilaria vermiculophylla. Methods Mol Biol. 2015;1308:159–172. doi:10.1007/978-1-4939-2684-8_10.
- Young DLW, Wiegand MD, Loadman NL, Collins SA, Ballevona AJ, Huebner JD. Effects of artificial ultraviolet-B radiation on growth and fatty acid composition of duckweed (Lemna minor). Freshw Biol. 2006;51(11):2029–2040. doi:10.1111/j.1365-2427.2006.01633.x.
- Lopez D, Casero D, Cokus SJ, Merchant SS, Pellegrini M. Algal functional annotation tool: a web-based analysis suite to functionally interpret large gene lists using integrated annotation and expression data. BMC Bioinformatics. 2011;12(1):282. doi:10.1186/1471-2105-12-282.
- Nozaki H. Origin and evolution of the genera Pleodorina and Volvox (Volvocales). Biologia (Bratisl). 2003;58:425–431.
- Kim T, Yang Q. Peroxisome-proliferator-activated receptors regulate redox signaling in the cardiovascular system. World J Cardiol. 2013;5(6):164. doi:10.4330/wjc.v5.i6.164.
- Michalik L, Wahli W PPARs mediate lipid signaling in inflammation and cancer. PPAR Res. 2008;2008.
- Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86(2):465–514. doi:10.1152/physrev.00025.2005.
- Richardson CE, Hennebelle M, Otoki Y, Zamora D, Yang J, Hammock BD, Taha AY. Lipidomic analysis of oxidized fatty acids in plant and algae oils. J Agric Food Chem. 2017;65(9):1941–1951. doi:10.1021/acs.jafc.6b05559.
- Marcos R, Izquierdo Y, Vellosillo T, Kulasekaran S, Cascón T, Hamberg M, Castresana C. 9-Lipoxygenase-derived oxylipins activate brassinosteroid signaling to promote cell wall-based defense and limit pathogen infection. Plant Physiol. 2015;169(3):2324–2334. doi:10.1104/pp.15.00992.
- Hamberg M, Sanz A, Rodriguez MJ, Calvo AP, Castresana C. Activation of the fatty acid α-dioxygenase pathway during bacterial infection of tobacco leaves FORMATION OF OXYLIPINS PROTECTING AGAINST CELL DEATH. J Biol Chem. 2003;278(51):51796–51805. doi:10.1074/jbc.M310514200.
- Logemann E, Hahlbrock K. Crosstalk among stress responses in plants: pathogen defense overrides UV protection through an inversely regulated ACE/ACE type of light-responsive gene promoter unit. Proc Natl Acad Sci. 2002;99(4):2428–2432. doi:10.1073/pnas.042692199.
- Yin H, Xu L, Porter NA. Free radical lipid peroxidation: mechanisms and analysis. Chem Rev. 2011;111(10):5944–5972. doi:10.1021/cr200084z.
- Wang X, Devaiah SP, Zhang W, Welti R. Signaling functions of phosphatidic acid. Prog Lipid Res. 2006;45(3):250–278. doi:10.1016/j.plipres.2006.01.005.
- Munnik T, Testerink C. Plant phospholipid signaling:“in a nutshell.”. J Lipid Res. 2009;50(Supplement):S260–S265. doi:10.1194/jlr.R800098-JLR200.
- Nakamura Y, Koizumi R, Shui G, Shimojima M, Wenk MR, Ito T, Ohta H. Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc Natl Acad Sci. 2009;106(49):20978–20983. doi:10.1073/pnas.0907173106.
- Gasulla F. vom Dorp K, Dombrink I, et al. The role of lipid metabolism in the acquisition of desiccation tolerance in C raterostigma plantagineum: a comparative approach. Plant J. 2013;75(5):726–741. doi:10.1111/tpj.12241.
- Moellering ER, Muthan B, Benning C. Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science. 2010;330(6001):226–228. doi:10.1126/science.1191803.
- Okazaki Y, Otsuki H, Narisawa T, Kobayashi M, Sawai S, Kamide Y, Kusano M, Aoki T, Hirai MY, Saito K. A new class of plant lipid is essential for protection against phosphorus depletion. Nat Commun. 2013;4:1510. doi:10.1038/ncomms2512.
- Duque L, Bravo K, Osorio E. A holistic anti-aging approach applied in selected cultivated medicinal plants: a view of photoprotection of the skin by different mechanisms. Ind Crops Prod. 2017;97:431–439. doi:10.1016/j.indcrop.2016.12.059.
- Abbott JA, Francklyn CS, Robey-Bond SM. Transfer RNA and human disease. Front Genet. 2014;5:158. doi:10.3389/fgene.2014.00158.
- Nandakumar J, Schwer B, Schaffrath R, Shuman S. RNA repair: an antidote to cytotoxic eukaryal RNA damage. Mol Cell. 2008;31(2):278–286. doi:10.1016/j.molcel.2008.05.019.
- Schwer B, Sawaya R, Ho CK, Shuman S. Portability and fidelity of RNA-repair systems. Proc Natl Acad Sci. 2004;101(9):2788–2793. doi:10.1073/pnas.0305859101.
- Katz A, Orellana O. Protein synthesis and the stress response. In: Biyani M, editor. Cell-free protein synthesis. IntechOpen; 2012.
- Dammeyer T, Frankenberg-Dinkel N. Insights into phycoerythrobilin biosynthesis point toward metabolic channeling. J Biol Chem. 2006;281(37):27081–27089. doi:10.1074/jbc.M605154200.
- Hsieh P, Pedersen JZ, Albertano P. Generation of reactive oxygen species upon red light exposure of cyanobacteria from Roman hypogea. Int Biodeterior Biodegradation. 2013;84:258–265. doi:10.1016/j.ibiod.2012.11.007.
- Hsieh P, Pedersen JZ, Bruno L. Photoinhibition of cyanobacteria and its application in cultural heritage conservation. Photochem Photobiol. 2014;90(3):533–543. doi:10.1111/php.12208.
- Hideg É, Jansen MAKK, Strid Å. UV-B exposure, ROS, and stress: inseparable companions or loosely linked associates? Trends Plant Sci. 2013;18(2):107–115. doi:10.1016/j.tplants.2012.09.003.
- Michaelian K, Simeonov A. Fundamental molecules of life are pigments which arose and evolved to dissipate the solar spectrum. Biogeosciences Discuss. 2015;12(3):2101–2160. doi:10.5194/bgd-12-2101-2015.
- Chung DW, PruŽinská A, Hörtensteiner S, Ort DR. The role of pheophorbide a oxygenase expression and activity in the canola green seed problem. Plant Physiol. 2006;142(1):88–97. doi:10.1104/pp.106.084483.
- Hirashima M, Tanaka R, Tanaka A. Light-independent cell death induced by accumulation of pheophorbide a in Arabidopsis thaliana. Plant Cell Physiol. 2009;50(4):719–729. doi:10.1093/pcp/pcp035.
- Lim PO, Kim HJ, Nam HG. Leaf senescence. Annu Rev Plant Biol. 2007;58(1):115–136. doi:10.1146/annurev.arplant.57.032905.105316.
- Leuendorf JE, Osorio S, Szewczyk A, Fernie AR, Hellmann H. Complex assembly and metabolic profiling of Arabidopsis thaliana plants overexpressing vitamin B6 biosynthesis proteins. Mol Plant. 2010;3(5):890–903. doi:10.1093/mp/ssq041.
- Mooney S, Hellmann H. Vitamin B6: killing two birds with one stone? Phytochemistry. 2010;71(5–6):495–501. doi:10.1016/j.phytochem.2009.12.015.
- Ehrenshaft M, Bilski P, Li MY, Chignell CF, Daub ME. A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc Natl Acad Sci. 1999;96(16):9374–9378. doi:10.1073/pnas.96.16.9374.
- Chen H, Xiong L. Pyridoxine is required for post-embryonic root development and tolerance to osmotic and oxidative stresses. Plant J. 2005;44(3):396–408. doi:10.1111/j.1365-313X.2005.02538.x.
- Titiz O, Tambasco-Studart M, Warzych E, Apel K, Amrhein N, Laloi C, Fitzpatrick TB. PDX1 is essential for vitamin B6 biosynthesis, development and stress tolerance in Arabidopsis. Plant J. 2006;48(6):933–946. doi:10.1111/j.1365-313X.2006.02928.x.
- Adams CR, Stamer KA, Miller JK, McNally JG, Kirk MM, Kirk DL. Patterns of organellar and nuclear inheritance among progeny of two geographically isolated strains of Volvox carteri. Curr Genet. 1990;18(2):141–153. doi:10.1007/BF00312602.
- Starr RC. Structure, reproduction and differentiation in Volvox carteri f. nagariensis Iyengar, strains HK 9 & 10. Arch Protistenkunde. 1969;111:204–222.
- Provasoli L, Pxntner IJ. Artificial media for fresh-water algae: problems and suggestions. In Tryon CA, & Hartman RT, editors. The Ecology of Algae University of Pittsburg Press, Pittsburg. 1960;84–96.
- Starr RC, Jaenicke L. Purification and characterization of the hormone initiating sexual morphogenesis in Volvox carteri f. nagariensis Iyengar. Proc Natl Acad Sci. 1974;71(4):1050–1054. doi:10.1073/pnas.71.4.1050.
- Andrews S FastQC: a quality control tool for high throughput sequence data. 2010.
- Bolger A, Lohse M, Usadel B. Trimmomatic: a flexi ble trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. Bioinformatics. 2014;30(15):1–7. doi:10.1093/bioinformatics/btu170.
- Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK, et al. Genomic analysis of organismal complexity in the multicellular green alga volvox carteri. Science. 2010;329(5988):223–226. doi:10.1126/science.1188800.
- Trapnell C, Pachter L, Salzberg S. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi:10.1093/bioinformatics/btp120.
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi:10.1093/bib/bbs017.
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Erratum: differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. 2014;9(10):2513. doi:10.1038/nprot.2012.016.
- Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI. A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci. 2005;102(50):18225–18230. doi:10.1073/pnas.0507187102.
- Favory J, Gruber H, Rizzini L, Oravecz A, Funk M, Albert A, Cloix C, Jenkins GI, Oakeley EJ, Seidlitz HK, et al. Interaction of COP1 and UVR8 regulates stress acclimation in Arabidopsis. 2009;(January): 591–601. doi:10.1038/emboj.2009.4.
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(D1):D1178–D1186. doi:10.1093/nar/gkr944.
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10(1):421. doi:10.1186/1471-2105-10-421.
