ABSTRACT
Cytokinins play an essential role in plant growth and development. A recent study showed that the cytokinin signaling pathway was conserved in the liverwort Marchantia polymorpha, and that it controlled gemma cup and rhizoid formation during thallus development. Here we show that the type-B response regulator, MpRRB, is mainly localized in the nucleus. Moreover, observations of thalli revealed that the distribution of air pores and the shape of the thallus margin are impaired in cytokinin-deficient lines and those defective in cytokinin signaling. This suggests that cytokinins regulate cell division and/or differentiation of precursor cells derived from the apical cell, thereby coordinating development of various organs produced on the thallus.
The phytohormone cytokinin is involved in a broad range of physiological events, such as cell division, organ growth and senescence.Citation1 Type-B response regulators (RRBs) are transcription factors that control cytokinin-responsive genes downstream of the two-component signaling pathway, which is mediated by CHASE domain-containing histidine kinase receptors and histidine-containing phosphotransfer proteins. Type-A response regulators (RRAs), one of the targets of RRBs, repress cytokinin signaling, thereby attenuating the response.Citation2,Citation3 Genetic analyses using Arabidopsis mutants have revealed that both types of RRs play an essential role in various developmental processes,Citation1 but how they precisely coordinate downstream events remains largely unknown.
The liverwort Marchantia polymorpha is a model basal land plant.Citation4,Citation5 The organization of the thalloid body derives from highly regulated division of the apical cell and its descendants at the thallus tip, which is called the notch. Arabidopsis possesses 10 and 11 genes encoding RRA and RRB, respectively. However, M. polymorpha only has one gene for each of RRA (MpRRA) and RRB (MpRRB).Citation6,Citation7 We recently reported that promoter activity of MpRRB was observed at the notch of young thalli and that transgenic plants defective in cytokinin signaling, e.g. Mprrb knockout lines, formed less or no gemma cup and more rhizoids than wild-type plants.Citation7 Transgenic lines overexpressing cytokinin oxidase (CKX), which inactivates cytokinins,Citation8 had less or no gemmiparous cells near the apical cell.Citation7 These findings indicate that cytokinins participate in the early process of gemma cup and rhizoid formation. However, the involvement of cytokinin signaling in generating other organs or tissues has yet to be uncovered.
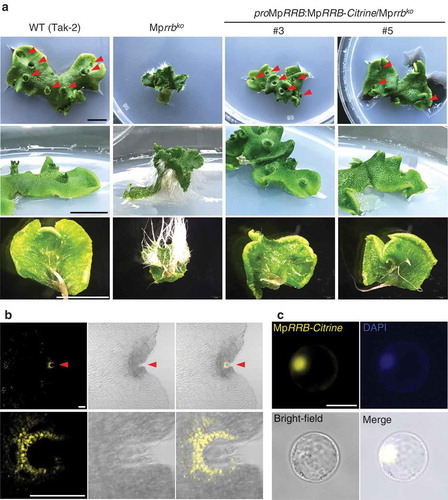
In Arabidopsis, RRBs are localized in the nucleus to exert transcriptional activity.Citation9,Citation10 To examine the subcellular localization of MpRRB, we fused a genomic fragment containing its promoter and coding regions to the gene for the fluorescent protein Citrine, and expressed the MpRRB-Citrine fusion protein in the Mprrb knockout line. As reported by Aki et al. (2019),Citation7 thalli with numerous rhizoids bent upward, and no gemma cup was formed in the Mprrb knockout line. These phenotypes were suppressed in the two knockout lines expressing MpRRB-Citrine (#3 and #5) (), indicating that MpRRB-Citrine is functional. To observe the localization of MpRRB-Citrine, gemmae were cultured for five days and fixed in PBS containing 4% paraformaldehyde at 4°C overnight, followed by clearing with the ClearSee solution for five days.Citation12 Citrine signals were detected at the apical notch (), upper), and a magnified view showed that a higher signal was observed in the nucleus while a significant level of fluorescence was also detected in the cytosol (), lower). We then transiently expressed MpRRB-Citrine in Arabidopsis protoplasts prepared from suspension cultured cells, and found that distinct Citrine signals were detected in the nucleus (). These results suggest that MpRRB is mainly localized in the nucleus, while it is possible that a small part of the protein is also present in the cytosol.
Figure 1. Subcellular localization of MpRRB. (a) Thalli of the Mprrb knockout line and the same line carrying proMpRRB:MpRRB-Citrine. The genomic fragment containing the promoter and the coding regions of MpRRB was introduced into the binary vector pMpGWB307,Citation11 which was then transformed into the Mprrb knockout line having an F1 background generated by crossing Tak-1 and Tak-2. Thallus tips were cultured for 15 days. Arrowheads indicate gemma cups. Bars represent 1 cm. (b) Expression and protein localization of Citrine-fused MpRRB at the notch. Gemmae were cultured for five days, and harvested thalli were fixed and cleared. Images were obtained with the confocal microscope (FV1000, Olympus). The lower panels are enlarged images of the notch region. Fluorescence images of Citrine signals (left); brightfield images (middle); merged images (right). Arrowheads indicate apical notches. Bars represent 100 µm. (c) Protein localization of Citrine-fused MpRRB in Arabidopsis protoplasts. MpRRB-Citrine was expressed under the cauliflower mosaic virus 35S promoter in Arabidopsis protoplasts prepared from suspension cultured cells. Fluorescence image of Citrine signal (upper left); DAPI-stained nuclei (upper right); bright-field image (lower left); merged image (lower right). Bar represents 20 µm.
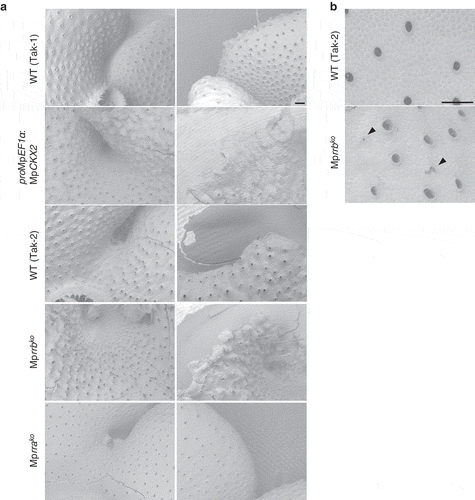
To understand the role of cytokinins in organ development in more detail, we observed thalli of MpCKX2-overexpressing plants and knockout lines for MpRRA and MpRRB with scanning electron microscopy. Observations of the dorsal side of thalli revealed that air pores were uniformly dispersed in wild-type plants but were irregularly distributed in the MpCKX2-overexpressing line and the Mprrb knockout line; there was extensive variation in the distance between pores (), left). In the Mprrb knockout line, we found air pores without a circular shape (), arrowheads). We could not observe any differences in the distribution and the shape of air pores between wild-type plants and the Mprra knockout line, in which cytokinin signaling was activated (), left). Around the notch region, the MpCKX2-overexpressing and Mprrb knockout lines displayed ectopic serrations on the thallus margin, which was never observed in wild-type plants (), right). Ectopic serrations in MpRRB knockdown plants also have been described by Flores-Sandoval et al. (2016).Citation13 We could not observe thallus margins of the Mprra knockout line, because they were buried in curled thalli (), right). Our results indicate that in addition to gemma cup and rhizoid formation,Citation7 cytokinin signaling controls the formation of air pores and thallus margins during thallus development.
Figure 2. Scanning electron micrographs of the MpCKX2-overexpressing line and the knockout lines of MpRRA and MpRRB. (a) Apical notch regions (left) and thallus margins (right). The MpCKX2-overexpressing line and the knockout lines of MpRRA and MpRRB are male and female lines, which have Tak-1 and F1 background, respectively. (b) Magnified images of the dorsal side of thalli. Thallus tips were cultured for 14 days and observed with a scanning electron microscope TM3000 (hitachi high technologies). Arrowheads indicate air pores without a circular shape. Bars represent 200 µm.
In M. polymorpha, all tissues in the thallus body derive from four merophytes (dorsal, ventral and two lateral merophytes) that are generated from the single apical cell at the notch.Citation5 Gemma cups originate from the dorsal merophyte, while rhizoids and air pores are produced from the ventral and dorsal sides of lateral merophytes, respectively. Although the origins of the thallus margin remain elusive, it is likely derived from lateral merophytes whose descendants undergo dorsiventral and/or coplanar cell division to form its smooth shape. While it remains unknown whether the observed defects in the MpCKX2-overexpressing and Mprrb knockout lines were due to impaired cell division or cell differentiation, our results suggest that cytokinin signaling regulates precursor cells derived from all merophytes, thereby enabling coordinated development of different organs during thallus growth. Further studies will be necessary to reveal the roles of cytokinins in cell division and/or differentiation during the early process of organ formation.
Abbreviations
| RR | = | response regulator |
| CKX | = | cytokinin oxidase |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Kieber JJ, Schaller GE. Cytokinins. Arabidopsis Book. 2014;12:1. doi:10.1199/tab.0168.
- To JPC, Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 2004;16:658–4. doi:10.1105/tpc.018978.
- To JPC, Deruère J, Maxwell BB, Morris VF, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ. Cytokinin regulates type-A Arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell. 2007;19:3901–3914. doi:10.1105/tpc.107.052662.
- Ishizaki K, Nishihama R, Yamato KT, Kohchi T. Molecular genetic tools and techniques for Marchantia polymorpha research. Plant Cell Physiol. 2016;57:262–270. doi:10.1093/pcp/pcv097.
- Shimamura M. Marchantia polymorpha: taxonomy, phylogeny and morphology of a model system. Plant Cell Physiol. 2016;57:230–256. doi:10.1093/pcp/pcv192.
- Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F, et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 2017;171:287–304. e15. doi:10.1016/j.cell.2017.09.030.
- Aki SS, Mikami T, Naramoto S, Nishihama R, Ishizaki K, Kojima M, Takebayashi Y, Sakakibara H, Kyozuka J, Kohchi T, et al. Cytokinin signaling is essential for organ formation in Marchantia polymorpha. Plant Cell Physiol. 2019;60:1842–1854. doi:10.1093/pcp/pcz100.
- Schmülling T, Werner T, Riefler M, Krupkova E, Bartrina Y, Manns I. Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J. Plant Res. 2003;116:241–252. doi:10.1007/s10265-003-0096-4.
- Mason MG, Li J, Mathews DE, Kieber JJ, Schaller GE. Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol. 2005;135:927–937. doi:10.1104/pp.103.038109.
- Xie M, Chen H, Huang L, O’Neil RC, Shokhirev MN, Ecker JR. A B-ARR-mediated cytokinin transcriptional network directs hormone cross-regulation and shoot development. Nat Commun. 2018;9:1604–1616. doi:10.1038/s41467-018-03921-6.
- Ishizaki K, Nishihama R, Ueda M, Inoue K, Ishida S, Nishimura Y, Shikanai T, Kohchi T, Ezura H. Development of gateway binary vector series with four different selection markers for the liverwort Marchantia polymorpha. PLoS One. 2015;10:e0138876. doi:10.1371/journal.pone.0138876.
- Kurihara D, Mizuta Y, Sato Y, Higashiyama T. ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development. 2015;142:4168–4179. doi:10.1242/dev.127613.
- Flores-Sandoval E, Dierschke T, Fisher TJ, Bowman JL. Efficient and inducible use of artificial microRNAs in Marchantia polymorpha. Plant Cell Physiol. 2016;57:281–290. doi:10.1093/pcp/pcv068.
