ABSTRACT
The plant trans-Golgi Network/Early Endosome (TGN/EE), as an organizer of vesicle trafficking, fulfills a crucial role for plant development and adaptation. Because it coordinates the transport of cell material along different routes, it is expected that a number of TGN/EE associated factors function in the rapid organization of post-Golgi trafficking to ensure that proteins reach their destination. The roles of Transport Protein Particle (TRAPP) complexes in the regulation of plant post-Golgi trafficking start to emerge. We previously demonstrated that the plant TRAPPIII complex is involved in maintenance of TGN organization and function and has a role in endocytic trafficking mediated by the SYP61 TGN/EE compartment. Here we show that attrappc11 mutants display accumulation of the plasma membrane resident proteins CESA6, BRI1 and PIP1;4 in aberrant intracellular compartments. This adds further insights into the functions of TRAPPIII as a regulators of post-Golgi/endosomal traffic.
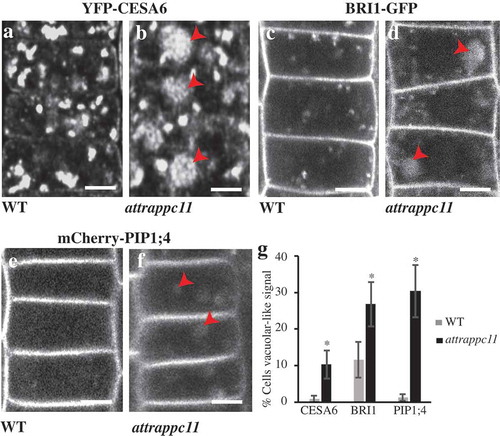
The plant SYP61 trans-Golgi Network/Early Endosome (TGN/EE) compartment is involved in trafficking of protein cargo between the Golgi and the plasma membrane (PM) along both the endocytic and exocytic routes.Citation1–Citation3 Plant post-Golgi trafficking is tightly controlled by a myriad of factors, including multi-subunit tethering complexes such as the EXOCYST, HOPS/CORVET and TRAPPs.Citation2,Citation4–Citation9 Only recently, the composition of TRAPP complexes and their roles in intracellular trafficking have been explored. So far, two TRAPP complexes, TRAPPII and TRAPPIII, have been found in plant cells.Citation4,Citation5 Both associate with the TGN/EE where they likely display compartmentalized functions.Citation4,Citation5,Citation10–Citation12 Previously, we showed that the plant TRAPPIII associates with the SYP61 TGN/EE population and that it has roles in the maintenance of the TGN/EE organization and function and the SYP61-mediated trafficking of the endocytic tracer FM4-64.Citation4 Fluorescently-tagged SYP61 is normally observed at TGN/EE and at the PM; however, in mutants of the TRAPPIII subunit AtTRAPPC11, a population of CFP-SYP61 displays aberrant localization to the tonoplast as confirmed using SNARF-1 vacuolar staining.Citation4 The latter raises the question whether an alternative pathway is activated in cells where the activity of TRAPPIII is inhibited. To investigate this, we examined the localization of three PM protein markers: Cellulose Synthase 6 (CESA6), Brassinosteroid Insensitive 1 (BRI1) and PM Intrinsic Protein 1;4 (PIP1;4) in root cells of attrappc11 mutants. CESAs are known cargoes of the SYP61 vesiclesCitation1 and evidence exists for SYP61-mediated trafficking of both BRI1 and PIP1;4.Citation3,Citation13 In the wild type background, fluorescently-tagged versions of all three proteins displayed the expected, previously reported subcellular localization pattern. Both YFP-CESA6 and BRI1-GFP localized to the PM and TGN/EE punctae,Citation14,Citation15 () while mCherry-PIP1;4 localized almost exclusively to the PMCitation16 ()). Interestingly, when expressed in the attrappc11 background, all three proteins frequently showed aberrant localization in intracellular compartments that resemble vacuoles (). This is consistent with the aberrant localization of SYP61 into the tonoplast in the mutant.Citation4 Such findings suggest the existence of a trafficking pathway that is activated, favored or released in the absence of a functional TRAPPIII complex, resulting in the accumulation of, at least, a set of PM proteins in aberrant intracellular “vacuole-like” compartments. Future analysis is necessary to identify the nature of these aberrant compartments using endomembrane markers. Interestingly, defective delivery of PM proteins has been reported also for mutants of the plant TRAPPII.Citation10,Citation11,Citation17 The latter, together with the observed intracellular accumulation of BRI1, PIP1;4 and CESA6 in attrappc11 mutants, indicates that both plant TRAPPs regulate the transport of PM proteins. Rerouting of PM proteins in attrappc11 mutants may point to the existence of trafficking regulation within the TGN/EE, ensuring that PM resident proteins reach their destination, with the involvement of TRAPPIII. A role for AtTRAPPC11 and TRAPPIII in such an intriguing mechanism will be the subject of future studies.
Figure 1. Localization of YFP-CESA6, BRI1-GFP and mCherry-PIP1;4 in root cells of attrappc11 mutants. (a, c and e) In the wild type background, the plasma membrane (PM) proteins CESA6 (a), BRI1 (c) and PIP1;4 (e) are observed at the PM and Golgi/TGN in root cells. (b, d and f) In the attrappc11 mutant background, aberrant localization (arrowheads) is frequently observed for the three proteins (b, d and f, respectively), in addition to PM and TGN/Golgi localization. (g) The graph shows the percentage of root cells where aberrant localization of YFP-CESA6, BRI1-GFP and mCherry-PIP1;4 is observed in seedlings of the WT and attrapc11 backgrounds. n > 10 seedlings, n > 20 cells per seedling. (*) P < .05, Student’s T test. Error bars represent standard errors. Scale bar = 5 μm.
Mutants of SYP61 are salt hypersensitive,Citation18 reflecting a role of the SYP61-mediated trafficking in the timely and selective delivery of PM proteins necessary for the stress response. Interestingly, a similar defect is observed in mutants of AtTRAPPC11.Citation4 We previously showed that AtTRAPPC11, and plausibly TRAPPIII, regulates SYP61-mediated post-Golgi trafficking.Citation4 Thus, it is tempting to speculate that defective trafficking of CESA6, BRI1, PIP1; and other PM residents critical the stress response,Citation19–Citation24 evidenced by their abnormal intracellular accumulation, is responsible for the salt hypersensitivity of attrappc11.
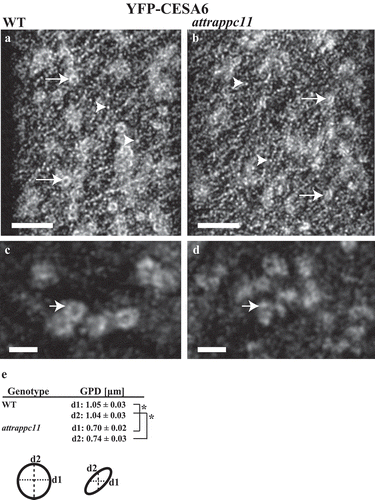
Cellulose synthase subunits (CESAs) are assembled into complexes (CSCs) that are transported by post-Golgi vesicles to the PM.Citation25–Citation28 In etiolated wild type hypocotyl cells, YFP-CESA6 particles associated with Golgi have the appearance of ring-shaped structures, while PM associated CESAs appear as smaller punctaeCitation14 (,,)). Interestingly, in attrappc11, the CESAs do not form the characteristic ring type but irregularly-shaped structures with a reduced diameter, when compared to the wild type (,,)). The assembly of CSC complexes is thought to be coordinated in the Golgi apparatus by the STELLO proteins.Citation29 Whether the altered appearance of CESA6 in attrappc11 mutants reflects a disrupted association of CSCs with Golgi stacks or a feedback mechanism of CSC regulation due to altered TRAPPIII function at the TGN/EE, are relevant questions to be explored.
Figure 2. The Golgi/TGN association of CESA6 complexes is affected in attrappc11 mutants. (a) In Col-0 wild type background (WT), CESA complexes (CSCs) associated with the Golgi appear as ring-like structure (arrows), while the smaller particles (arrowheads) represent CSCs at the PM. Z-projections. Scale bar= 5 μm. (b) The large ring structures are lost in the attrappc11 mutant background. (c, d) CSCs observed using Stimulated Emission Depletion (STED) super-resolution microscopy show an irregular shape in the attrappc11 mutant background (d) compared to WT (c). Z-projections. Scale bar= 2 μm. (e) The CSC particles arranged in ring shape structures are significantly reduced in diameter in the attrappc11 mutants, compared with WT. (*) P < .001, Student’s T test. GPD: Golgi/TGN particle diameter. d1: horizontal diameter. d2: vertical diameter. Error values represent standard errors. A cartoon with two hypothetical ring structured particles is provided to illustrate d1 and d2.
In conclusion, the accumulation of CESA6, BRI1 and the aquaporin PIP1;4 in intracellular compartments of mutants of the plant TRAPPIII subunit AtTRAPPC11 points toward a regulatory role of the complex in TGN-mediated trafficking of PM resident proteins. Mutants of the two plant TRAPP complexes identified so far, TRAPPII and TRAPPIII, display defects in PM protein localization,Citation4,Citation12 which opens exciting questions such as how instrumental TGN associated TRAPPs are for the plant endomembrane system’s plasticity and to which extent their post-Golgi trafficking functions are compartmentalized.
1. Materials and methods
1.1. Plant material and growth
Arabidopsis seedlings of the Columbia ecotype (Col-0) were used in this study. The T-DNA insertional mutant line of AtTRAPPC11, SALK119008 (attrappc11-2) was obtained from the Arabidopsis Biological Resources Center (ABRC) (http://www.arabidopsis.org;Citation30) and genetically characterized in a previous studyCitation4. The following Arabidopsis lines have been described previously: UBQ10pro:mCherry-PIP1;4 (WAVE 138RCitation16), CESA6::YFP-CESA6Citation14 and BRI1::BRI1-GFP.Citation31 Genetic crosses of the above-mentioned PM marker lines with attrappc11-2 mutants were established in this study. Seeds were sterilized using 30% (v/v) sodium chlorate in ethanol (absolute) with 0.06% (v/v) of Triton X-100 (Sigma-Aldrich). Seeds were plated on 0.25 Murashige and Skoog medium (1.15 g L−1 Murashige and Skoog minimal organics medium, 10 g L−1 sucrose, 5 g L−1 Phytagel (Sigma-Aldrich), and cold vernalized for 48 h at 4°C in the dark, after which plates were transferred to a plant growth chamber for seedling growth. Plants were grown in temperature- and photoperiod-controlled environments, set to long-day (16-h-light/8-h-dark cycle) conditions, using fluorescent light (at 100 to 150 mmol quanta photosynthetically active radiation [PAR] m–2 s–1) at 22 to 24°C.
1.2. Light microscopy and image analysis
A Leica SP8 confocal microscope was used for localization studies of mCherry-PIP1;4, BRI1-GFP and YFP-CESA6. Fluorescence signals of mCherry (excitation 587 nm, emission 598 to 684 nm), GFP (excitation 488 nm, emission 493 to 549 nm) and YFP (excitation 513 nm, emission 518 to 582 nm) were collected with 63× (oil), and 100× (oil) objectives. Stimulated Emission Depletion (STED) was used for imaging of YFP-CESA6. Image analysis was performed using LAS AF lite and Image J. Data represent images from more than ten independent seedlings. The diameter of CESA6-labeled Golgi/TGN compartments was measured using the line tool of the ImageJ software.Citation32
1.3. Statistical analysis
P-values were calculated with a two-tailed Student’s t test (R Development Core Team, 2006Citation33).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Drakakaki G, van de Ven W, Pan S, Miao Y, Wang J, Keinath NF, Weatherly B, Jiang L, Schumacher K, Hicks G, et al. Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res. 2012;22(2):1–4. doi:10.1038/cr.2011.129.
- Rosquete MR, Davis DJ, Drakakaki G. The plant trans-Golgi network: not just a matter of distinction. Plant Physiol. 2018;176(1):187–198. doi:10.1104/pp.17.01239.
- Robert S, Chary SN, Drakakaki G, Li S, Yang Z, Raikhel NV, Hicks GR. Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc Natl Acad Sci U S A. 2008;105(24):8464–8469. doi:10.1073/pnas.0711650105.
- Rosquete MR, Worden N, Ren G, Sinclair RM, Pfleger S, Salemi M, Phinney BS, Domozych D, Wilkop T, Drakakaki G. AtTRAPPC11/ROG2: a role for TRAPPs in maintenance of the plant trans-Golgi network/early endosome organization and function. Plant Cell. 2019;31(8):1879–1898. doi:10.1105/tpc.19.00110.
- Kalde M, Elliott L, Ravikumar R, Rybak K, Altmann M, Klaeger S, Wiese C, Abele M, Al B, Kalbfuß N, et al. Interactions between Transport Protein Particle (TRAPP) complexes and Rab GTPases in Arabidopsis. Plant J. 2019. doi:10.1111/tpj.14442.
- Vukasinovic N, Zarsky V. Tethering complexes in the Arabidopsis endomembrane system. Front Cell Dev Biol. 2016;4:46. doi:10.3389/fcell.2016.00046.
- Zarsky V, Kulich I, Fendrych M, Pečenková T. Exocyst complexes multiple functions in plant cells secretory pathways. Curr Opin Plant Biol. 2013;16(6):726–733. doi:10.1016/j.pbi.2013.10.013.
- Ravikumar R, Steiner A, Assaad FF. Multisubunit tethering complexes in higher plants. Curr Opin Plant Biol. 2017;40:97–105. doi:10.1016/j.pbi.2017.08.009.
- Takemoto K, Ebine K, Askani JC, Krüger F, Gonzalez ZA, Ito E, Goh T, Schumacher K, Nakano A, Ueda T. Distinct sets of tethering complexes, SNARE complexes, and Rab GTPases mediate membrane fusion at the vacuole in Arabidopsis. Proc Natl Acad Sci U S A. 2018;115(10):E2457–E2466. E2457-e2466. doi:10.1073/pnas.1717839115.
- Qi X, Kaneda M, Chen J, Geitmann A, Zheng H. A specific role for Arabidopsis TRAPPII in post-Golgi trafficking that is crucial for cytokinesis and cell polarity. Plant J. 2011;68(2):234–248. doi:10.1111/j.1365-313X.2011.04681.x.
- Qi X, Zheng H. Arabidopsis TRAPPII is functionally linked to Rab-A, but not Rab-D in polar protein trafficking in trans-Golgi network. Plant Signal Behav. 2011;6(11):1679–1683. doi:10.4161/psb.6.11.17915.
- Ravikumar R, Kalbfuss N, Gendre D, Steiner A, Altmann M, Altmann S, Rybak K, Edelmann H, Stephan F, Lampe M, et al. Independent yet overlapping pathways ensure the robustness and responsiveness of trans-Golgi network functions in Arabidopsis. Development. 2018;145(21). pii: dev169201. doi:10.1242/dev.169201.
- Hachez C, Laloux T, Reinhardt H, Cavez D, Degand H, Grefen C, De Rycke R, Inzé D, Blatt MR, Russinova E, et al. Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2;7 to modulate the cell membrane water permeability. Plant Cell. 2014;26(7):3132–3147. doi:10.1105/tpc.114.127159.
- Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312(5779):1491–1495. doi:10.1126/science.1126551.
- Geldner N, Hyman DL, Wang X, Schumacher K, Chory J. Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 2007;21(13):1598–1602. doi:10.1101/gad.1561307.
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof Y-D, Chory J. Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 2009;59(1):169–178. doi:10.1111/j.1365-313X.2009.03851.x.
- Rybak K, Steiner A, Synek L, Klaeger S, Kulich I, Facher E, Wanner G, Kuster B, Zarsky V, Persson S, et al. Plant cytokinesis is orchestrated by the sequential action of the TRAPPII and exocyst tethering complexes. Dev Cell. 2014;29(5):607–620. doi:10.1016/j.devcel.2014.04.029.
- Zhu JH, Gong Z, Zhang C, Song C-P, Damsz B, Inan G, Koiwa H, Zhu J-K, Hasegawa PM, Bressan RA. OSM1/SYP61: A syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. Plant Cell. 2002;14(12):3009–3028. doi:10.1105/tpc.006981.
- Planas-Riverola A, Gupta A, Betegón-Putze I, Bosch N, Ibañes M, Caño-Delgado AI. Brassinosteroid signaling in plant development and adaptation to stress. Development. 2019;146(5):dev151894. doi:10.1242/dev.151894.
- Kesten C, Wallmann A, Schneider R, McFarlane HE, Diehl A, Khan GA, van Rossum B-J, Lampugnani ER, Szymanski WG, Cremer N, et al. The companion of cellulose synthase 1 confers salt tolerance through a Tau-like mechanism in plants. Nat Commun. 2019;10(1):857. doi:10.1038/s41467-019-08780-3.
- Zhang -S-S, Sun L, Dong X, Lu S-J, Tian W, Liu J-X. Cellulose synthesis genes CESA6 and CSI1 are important for salt stress tolerance in Arabidopsis. J Integr Plant Biol. 2016;58(7):623–626. doi:10.1111/jipb.12442.
- Kapilan R, Vaziri M, Zwiazek JJ. Regulation of aquaporins in plants under stress. Biol Res. 2018;51(1):4. doi:10.1186/s40659-018-0176-5.
- Luu DT, Martinière A, Sorieul M, Runions J, Maurel C. Fluorescence recovery after photobleaching reveals high cycling dynamics of plasma membrane aquaporins in Arabidopsis roots under salt stress. Plant J. 2012;69(5):894–905. doi:10.1111/j.1365-313X.2011.04841.x.
- Endler A, Schneider R, Kesten C, Lampugnani ER, Persson S. The cellulose synthase companion proteins act non-redundantly with CELLULOSE SYNTHASE INTERACTING1/POM2 and CELLULOSE SYNTHASE 6. Plant Signal Behav. 2016;11(4):e1135281. doi:10.1080/15592324.2015.1135281.
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AMC, Ehrhardt DW. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol. 2009;11(7):797–806. doi:10.1038/ncb1886.
- Worden N, Park E, Drakakaki G. Trans-Golgi network-an intersection of trafficking cell wall components(f). J Integr Plant Biol. 2012;54(11):875–886. doi:10.1111/j.1744-7909.2012.01179.x.
- Sinclair R, Rosquete MR, Drakakaki G. Post-Golgi trafficking and transport of cell wall components. Front Plant Sci. 2018;9:1784. doi:10.3389/fpls.2018.01784.
- Lampugnani ER, Khan GA, Somssich M, Persson S. Building a plant cell wall at a glance. J Cell Sci. 2018;131(2). pii: jcs207373. doi 10.1242/jcs.207373
- Zhang Y, Cui X, Zhang X, Cheng L, Liu Y, Yang Y, Fan P, Wang Q, Lin Y, Zhang J, et al. Golgi-localized STELLO proteins regulate the assembly and trafficking of cellulose synthase complexes in Arabidopsis. Nat Commun. 2016;7:11656. doi:10.1038/ncomms11996.
- Alonso JM, Strepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R et al. Genome-wide insertional mutagenesis of arabidopsis thaliana. Science. 2003; 301(5633):653-657. doi:10.1126/science.1086391
- Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123(4):1247–1256. doi:10.1104/pp.123.4.1247.
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675.
- R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2006. http://www.R-project.org.
