ABSTRACT
Plasmopara viticola, the causal oomycete of grapevine downy mildew disease, secrets a series of RXLR effectors to manipulate host immunity. In this study, we characterized the role of a RXLR effector of P. viticola, PvRXLR159, in plant–microbe interaction. Transcription of PvRXLR159 in P. viticola was induced in the early stage of infection in grapevine (Vitis vinifera ‘Thomson Seedless’). Further results revealed that PvRXLR159 contains a functional signal peptide and its C terminus was essential to inhibit cell death by elicitors, INF1 and BAX, in Nicotiana benthamiana. Transient expression of PvRXLR159 suppressed N. benthamiana resistance to a pathogenic oomycete, Phytophthora capsici. Taken together, we propose that PvRXLR159 is induced and secreted by P. viticola to suppress host resistance.
Introduction
Plants have evolved a two-layered innate immune system against pathogen infection. The first layer is termed as pattern-triggered immunity (PTI), which is triggered by the recognition via pattern recognition receptors (PRRs) of conserved molecules from microbes called pathogen/microbe-associated molecular patterns (PAMPs/MAMPs), such as bacterial flagellin peptides and an elicitin INF1 from oomycete. As a ubiquitous defense response in plants, PTI generally includes a series of rapid responses such as a burst of reactive oxygen species (ROS), callose deposition, activation of mitogen-activated protein kinases (MAPKs) and defense genes expression.Citation1–Citation4 On the other hand, most pathogens like bacteria, fungi, and oomycetes secrete effectors to suppress PTI for successful infection. As a counter measure, resistant plants recognize these effectors via resistance (R) proteins triggering the second layer of plant defense termed as effector-triggered immunity (ETI), limiting the proliferation of pathogens.Citation5
RXLR effectors (for Arg, any amino acid, Leu and Arg) and CRNs effectors (crinkling and necrosis induced protein) are two important classes of effectors in pathogenic oomycetes, and both of them contain modular structures, including N-terminal signal peptide responsible for transporting effector proteins into cells followed by some conserved motifs, like RXLR, QXLR, LFLAK and CHXC (X stands for any amino acid).Citation5–Citation10 So far, more than 500 putative RXLR and CRN effectors have been predicted by bioinformatics analysis.Citation6,Citation11–Citation13 A major function of those effectors is to suppress plant resistance.Citation14 It is known that C-terminal domains of RXLR effectors is generally important for their function in manipulating plant immunity.Citation15
Plasmopara viticola is an obligate biotrophic oomycete that causes devastating downy mildew disease of grapevine, resulting in tremendous economic loss in the grape and wine industry worldwide.Citation16 So far, more than 100 candidate RXLR effectors have been predicted from P. viticola.Citation17–Citation22 A comprehensive functional study of 83 PvRXLR effectors has found that 52 of them including PvRXLR159 completely suppressed cell death induced by elicitors, INF1 and BAX, when expressed in Nicotiana benthamiana, suggesting that they are potential suppressors in manipulating plant immunity.Citation23 Our recent studies revealed that PvRXLR131 suppresses host resistance by targeting BRI1 kinase inhibitor 1.Citation24 In this study, we characterized the role of PvRXLR159 in plant–microbe interaction.
Materials and methods
Plant materials and growth conditions
The grapevine (V. vinifera ‘Thompson Seedless’) and N. benthamiana used in this study were grown in a green house at 25ºC with a photoperiod of 14 h light/10 h darkness.
Vector construction
The oligonucleotides used for plasmid construction in this study are documented in the Supporting Information (Table S1). The signal peptide sequence of PvRXLR159 was predicted using the online site SingalP4.0 Server (http://www.cbs.dtu.dk/services/SignalP-4.0/). EcoRI and XhoI restriction enzymes were subsequently used to insert the signal peptide sequences into the pSUC2 vector. For the deletion experiment of suppressing cell death in N. benthamiana, different truncated sequences of PvRXLR159 were inserted into vector pGR106 using the SalI and ClaI restriction enzymes. For Phytophothora capsici infection assay, the sequence encoding PvRXLR159 without signal peptide was integrated into the vector PBI121-EGFP using BamHI and SpeI restriction enzymes.
Functional verification of signal peptide
Functional validation of the predicted signal peptide of PvRXLR159 was conducted with signal sequence trap (SST) assay.Citation25,Citation26 The pSUC2 vector used in this experiment contains the coding sequence of a truncated invertase, SUC2, without the initial methionine and signal peptide. The sequences of signal peptides of Avr1b and PvRXLR159, and the first 25 amino acids of Mg87 were fused in frame to the invertase gene in the pSUC2 vector, respectively. The recombinant plasmids were then transformed into invertase secretion-deficient yeast strain YTK12 by lithium acetate-mediated transformation,Citation27 and then the suspension was inoculated on CMD-W medium to screen the positive transformants. The positive yeast colonies were transferred to YPRAA medium and 0.1% 2, 3, 5-triphenyltetrazolium chloride (TTC) solution to verify the secretion of invertase.
Agrobacterium tumefacien-mediated transient expression in N. benthamiana
Plasmids were transformed into A. tumefacien strain GV3101 via electroporation, and then incubated in LB medium supplemented with appropriate antibiotics for 2 days.Citation28 The cells were harvested by centrifugation at 4000 × g for 5 min and resuspended in 10 mM MgCl2 to adjust OD600 to 0.4 for cell death assay and 0.6 for other experiments. Finally, the suspension was infiltrated into N. benthamiana leaves from the abaxial side using needleless syringes for expression.Citation29
Pathogen infection assay
Infection of P. viticola ‘JL-7-2ʹ was performed according to previous studies.Citation20,Citation24 The susceptible grapevine (V. vinifera ‘Thompson Seedless’) leaf discs were inoculated with 30 µL spore suspension with a final concentration of 1 × 105 spores/mL on the abaxial side, and then incubated in an incubator (18ºC, 11 h light/13 h dark, 66% relative humidity). Phenotypic monitoring and sampling for RNA extraction were performed at indicated times. For P. capsici infection assay, P. capsici was grown on oatmeal agar medium (30 g oat, 20 g agar, 1 L water) for 4–5 days in the dark at 25ºC. Agar discs with a diameter of 7 mm harboring P. capsici mycelium were put on detached leaves of N. benthamiana for infection.
DNA and RNA extraction and real-time quantitative PCR analysis
DNA and RNA from plant tissues were extracted using CTAB methodCitation30 and a plant RNA kit (Omega, Norcross, GA, USA), respectively. cDNA was synthesized using One-Step gDNA Removal and cDNA Synthesis Super Mix kit (Trans Gen Biotech). qPCR was performed using SYBR Green Fast qPCR Mix and StepOne Real-Time PCR system (Bio-rad, Hercules, CA, USA). Primers for qPCR were listed in Table S2.
Protein extraction and western blot analysis
Protein of N. benthamiana was extracted according to our previous studies.Citation23 The plant tissues were ground into powder with liquid nitrogen, and then subjected to protein extraction in lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% (v/v) Triton X-100, 1 mM DTT, 1 mM PMSF, 1× protease inhibitor cocktail, 5 µM MG132). Equal amounts of protein were separated by SDS-PAGE and then transferred to nitrocellulose membrane. The membrane was blocked with 5% skim milk and then incubated with anti-GFP as primary antibody for 1 h, followed by incubation with secondary antibody for 1 h. The signal was detected by Super Signal West Femto ECL kit (Thermo Scientific, Rockford, IL, USA).
Results
PvRXLR159 transcription is induced in the early stage of infection
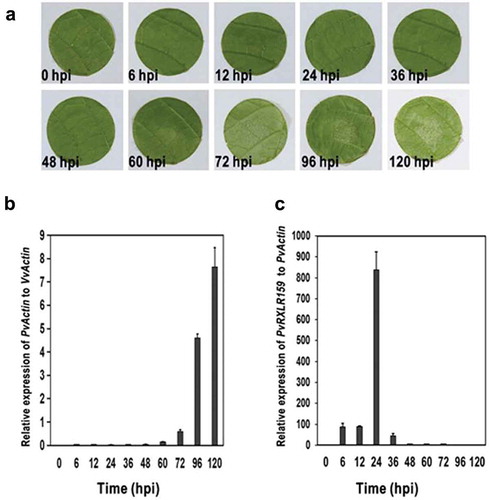
We investigated the expression pattern of PvRXLR159 in P. viticola during infection in grapevine by qRT-PCR. The growth of P. viticola on leaf discs was slow during the period from 0 to 60 h after inoculation, and then increased dramatically at later growth stages (). The abundance of PvRXLR159 transcripts peaked at around 24-h post-inoculation (hpi) and then dropped rapidly (). These results indicate that PvRXLR159 is induced in the early stage of infection, which may contribute to the growth of P. viticola in grapevine. Note that we could not detect transcripts of P. viticola at 0 hpi due to the very low abundance of P. viticola in leaf discs.
Figure 1. PvRXLR159 is induced in the early stage of infection.
A, Representative photos of grapevine leaf discs inoculated with P. viticola. B, The growth of P. viticola in grapevine. The growth of P. viticola was monitored by qPCR and evaluated as the relative quantity of PvActin (P. viticola Actin) to VvActin (V. vinifera Actin). C, Transcription of PvRXLR159 in P. viticola during infection. Transcription levels of PvRXLR159 were monitored by qPCR and evaluated as the relative quantity of PvRXLR159 to PvActin. Error bars represent the standard errors from three biological replicates. hpi, hours post-inoculation.
PvRXLR159 is an effector containing functional signal peptide
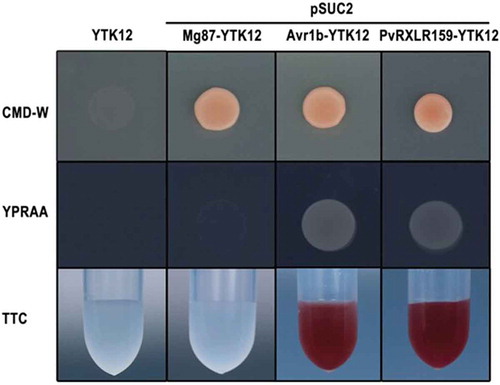
Previous studies have shown that many RXLR effectors have N-terminal signal peptides composed of hydrophobic amino acids and are secreted by oomycetes.Citation6,Citation31,Citation32 We used singalP4.0 to predict the signal peptide region of PvRXLR159 and found that the first 21 amino acids at the N terminus of PvRXLR159 are a potential signal peptide region with a high probability of 0.76. Hence, we performed the SST assay to validate whether the N-terminal signal peptide of PvRXLR159 has secretory function.Citation26,Citation33 The signal peptide sequences of Avr1b as a positive control and PvRXLR159, and the sequence coding the first 25 amino acids of Mg87 as a negative control were fused in frame to the invertase gene in the pSUC2 vector, which were transformed into the invertase secretion-deficient yeast strain YTK12.Citation34,Citation35 YTK12 carrying pSUC2-Avr1b and pSUC2-PvRXLR159 were able to grow on YPRAA medium with raffinose as the sole carbon source and reduce the dye TTC to the red-colored insoluble TTF, whereas the YTK12 strain without transformation or carrying pSUC2-Mg87 was not (), indicating that signal peptide of PvRXLR159 is functional.
Figure 2. PvRXLR159 is an effector containing functional signal peptide.
Functional validation of the signal peptide of PvRXLR159 was carried out by using signal sequence trap assay. The signal peptide sequences of PvRXLR159 and Avr1b as a positive control, and the sequence of the first 25 amino acids of Mg87 as a negative control were fused in frame to the invertase gene in the pSUC2 vector, which were transformed into yeast YTK12 strain. CMD-W medium were used to screen positive transformants. YPRAA medium and TTC reducing assay were used to test invertase secretion. Only clones having a functional signal peptide can grow on YPRAA medium and reduce TTC to red formazan.
C terminus of PvRXLR159 is essential for its function
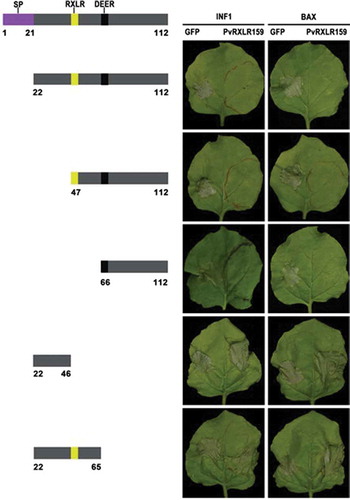
Our previous studies have shown that PvRXLR159 suppresses INF1-and BAX-triggered cell death when transiently expressed in N. benthamiana.Citation23 Based on this phenotype, we constructed several deletion mutants of PvRXLR159 to investigate which domain of PvRXLR159 is important for its function. The deletion mutants containing 66 to 112 C-terminal sequence of PvRXLR159 retained the ability to suppress INF1-and BAX-triggered cell death, whereas the mutants having N-terminal sequences did not (). These results indicate that the C terminus of PvRXLR159 is essential for blocking cell death triggered by INF1 and BAX.
Figure 3. C terminus of PvRXLR159 is essential for its function.
Deletion mutants of PvRXLR159 were expressed in N. benthamiana leaves for 24 h, followed by expression of the elicitors INF1 and BAX to induce cell death. Left panel, a schematic diagram of the different deletion constructs. Right panel, typical symptoms of N. benthamiana taken at 5 days after expression of INF1 and BAX. The experiments were repeated at least three times with similar results.
PvRXLR159 suppresses N. benthamiana resistance against phytophthora capsici
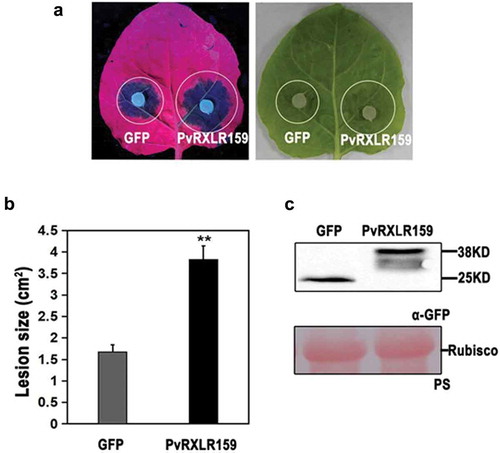
Since genetic modification of P. viticola and grapevine are both challenging, we investigated whether PvRXLR159 manipulates plant immunity against pathogenic oomycete using the N. benthamiana-P. capsici pathosystem instead. As shown in , expression of PvRXLR159 in N. benthamiana significantly increased the lesion size induced by P. capsici infection, indicating that infiltration of PvRXLR159 in N. benthamiana enhances susceptibility to P. capsici.
Figure 4. PvRXLR159 suppresses N. benthamiana resistance to P. capsici.
PvRXLR159-GFP and GFP were transiently expressed in N. benthamiana. Leaves were detached at 48 h, followed by inoculation with P. capsici. Images of typical symptoms were taken at 3 days after inoculated with P. capsici as shown in A. Quantification of lesion size in A was shown in B. Error bars represent the standard errors from three biological replicates. Asterisks represent significant differences from GFP (**P < .01, Student’s t-test). Expression of PvRXLR159-GFP was checked by western blot as shown in C. PS, Ponceau S staining.
Conclusions
In the present study, we functionally characterized PvRXLR159 with the following findings: 1) transcription of PvRXLR159 in P. viticola is induced during infection; 2) PvRXLR159 has a functional signal peptide; 3) C terminus is essential for its inhibition of INF1- and BAX-induced cell death in N. benthamiana; 4) PvRXLR159 suppresses N. benthamiana resistance against P. capsici. In addition, expression of PvRXLR159 also suppressed Arabidopsis resistance to a pathogenic bacterium, Pseudomonas syringae pv. tomato DC3000 (data not shown), suggesting that the suppression of host resistance by PvRXLR159 is a conserved phenotype in plants. Based on these results, we propose that PvRXLR159 is induced and secreted by P. viticola to suppress host resistance. Since PvRXLR159 is localized in both nucleus and cytoplasm when expressed in plants,Citation23 it may have different targets in these compartments. Research is undergoing to identify the targets of PvRXLR159 to shed light on the detail mechanism behind its interference effect on host resistance.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Supplemental Material
Download MS Word (14.3 KB)Supplemental Material
Download MS Word (15.7 KB)Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Macho AP, Zipfel C. Plant PRRs and the activation of innate immune signaling. Mol Cell. 2014;54(2):1–7. doi:10.1016/j.molcel.2014.03.028.
- Pitzschke A, Schikora A, Hirt H. MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol. 2009;12(4):421–426. doi:10.1016/j.pbi.2009.06.008.
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi:10.1146/annurev.arplant.57.032905.105346.
- Couto D, Zipfel C. Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol. 2016;16(9):537–552. doi:10.1038/nri.2016.77.
- Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11(8):539–548. doi:10.1038/nrg2812.
- Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461(7262):393–398. doi:10.1038/nature08358.
- Morgan W, Kamoun S. RXLR effectors of plant pathogenic oomycetes. Curr Opin Microbiol. 2007;10(4):332–338. doi:10.1016/j.mib.2007.04.005.
- Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, van West P, Chapman S, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450(7166):115–118. doi:10.1038/nature06203.
- Kamoun S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev Phytopathol. 2006;44:41–60. doi:10.1146/annurev.phyto.44.070505.143436.
- Liu L, Xu L, Jia Q, Pan R, Oelmüller R, Zhang W, Wu C. Arms race: diverse effector proteins with conserved motifs. Plant Signal Behav. 2019;14:2. doi:10.1080/15592324.2018.1557008.
- Baxter L, Tripathy S, Ishaque N, Boot N, Cabral A, Kemen E, Thines M, Ah-Fong A, Anderson R, Badejoko W, et al. Signatures of adaptation to obligate biotrophy in the hyaloperonospora arabidopsidis genome. Science. 2010;330(6010):1549–1551. doi:10.1126/science.1195203.
- Sharma R, Xia X, Cano LM, Evangelisti E, Kemen E, Judelson H, Oome S, Sambles C, van Den Hoogen DJ, Kitner M, et al. Genome analyses of the sunflower pathogen Plasmopara halstedii provide insights into effector evolution in downy mildews and Phytophthora. BMC Genomics. 2015;16:741. doi:10.1186/s12864-015-1904-7.
- Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313(5791):1261–1266. doi:10.1126/science.1128796.
- Wang Y, Tyler BM, Defense WY. Counterdefense during plant-pathogenic oomycete infection. Annu Rev Microbiol. 2019;73(1):667–696. doi:10.1146/annurev-micro-020518-120022.
- Oh S-K, Kamoun S, Choi D. Oomycetes RXLR effectors function as both activator and suppressor of plant immunity. Plant Pathol J. 2010;26(3):209–215. doi:10.5423/ppj.2010.26.3.209.
- Gessler C, Pertot I, Perazzolli M. Plasmopara viticola: a review of knowledge on downy mildew of grapevine and effective disease management. Phytopathol Mediterr. 2011;50(1):3–44. doi:10.1111/j.1365-3180.2010.00835.x.
- Brilli M, Asquini E, Moser M, Bianchedi PL, Perazzolli M, Si-Ammour A. A multi-omics study of the grapevine-downy mildew (Plasmopara viticola) pathosystem unveils a complex protein coding- and noncoding-based arms race during infection. Sci Rep. 2018;8:757. doi:10.1038/s41598-018-19158-8.
- Mestre P, Carrere S, Gouzy J, Piron M-C, Tourvieille de Labrouhe D, Vincourt P, Delmotte F, Godiard L. Comparative analysis of expressed CRN and RXLR effectors from two Plasmopara species causing grapevine and sunflower downy mildew. Plant Pathol. 2016;65(5):767–781. doi:10.1111/ppa.12469.
- Mestre P, Piron M-C MD. Identification of effector genes from the phytopathogenic Oomycete Plasmopara viticola through the analysis of gene expression in germinated zoospores. Fungal Biol. 2012;116(7):825–835. doi:10.1016/j.funbio.2012.04.016.
- Xiang J, Li X, Wu J, Yin L, Zhang Y, Lu J. Studying the mechanism of Plasmopara viticola RxLR effectors on suppressing plant immunity. Front Microbiol. 2016;7:709. doi:10.3389/fmicb.2016.00709.
- Yin L, An Y, Qu J, Li X, Zhang Y, Dry I, Wu H, Lu J. Genome sequence of Plasmopara viticola and insight into the pathogenic mechanism. Sci Rep. 2017;7:46553. doi:10.1038/srep46553.
- Yin L, Li X, Xiang J, Qu J, Zhang Y, Dry IB, Lu J, et al. Characterization of the secretome of Plasmopara viticola by de novo transcriptome analysis. Physiol Mol Plant Pathol. 2015;91:1–10. doi:10.1016/j.pmpp.2015.05.002.
- Liu Y, Lan X, Song S, Yin L, Dry IB, Qu J, Xiang J, Lu J. In planta functional analysis and subcellular localization of the Oomycete Pathogen Plasmopara viticola candidate RXLR effector repertoire. Front Plant Sci. 2018;9:286. doi:10.3389/fpls.2018.00286.
- Lan X, Liu Y, Song S, Yin L, Xiang J, Qu J, Lu J. Plasmopara viticola effector PvRXLR131 suppresses plant immunity by targeting plant receptor-like kinase inhibitor BKI1. Mol Plant Pathol. 2019;20(6):765–783. doi:10.1111/mpp.12790.
- Klein RD, Gu Q, Goddard A, Rosenthal A. Selection for genes encoding secreted proteins and receptors. Proc Natl Acad Sci U S A. 1996;93(14):7108–7113. doi:10.1073/pnas.93.14.7108.
- Oh S-K, Young C, Lee M, Oliva R, Bozkurt TO, Cano LM, Win J, Bos JIB, Liu H-Y, van Damme M, et al. In planta expression screens of phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell. 2009;21(9):2928–2947. doi:10.1105/tpc.109.068247.
- Gietz RD, Graham KC, Litchfield DW. Interactions between the subunits of casein kinase II. J Biol Chem. 1995;270(22):13017–13021. doi:10.1074/jbc.270.22.13017.
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol. 2000;42(6):819–832. doi:10.1023/a:1006496308160.
- Meng Y, Zhang Q, Zhang M, Gu B, Huang G, Wang Q, Shan W. The protein disulfide isomerase 1 of Phytophthora parasitica (PpPDI1) is associated with the haustoria-like structures and contributes to plant infection. Front Plant Sci. 2015;6:632. doi:10.3389/fpls.2015.00632.
- Samuel TO, Ebabhi AM, Adekunle AA. Identification of some human pathogenic fungi using four DNA extraction methods. J Appl Sci Environ Manage. 2017;21(6):1079–1084. doi:10.4314/jasem.v21i6.14.
- Dou D, Kale SD, Wang X, Jiang RHY, Bruce NA, Arredondo FD, Zhang X, Tyler BM. RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell. 2008;20(7):1930–1947. doi:10.1105/tpc.107.056093.
- Fawke S, Doumane M, Schornack S. Oomycete interactions with plants: infection strategies and resistance principles. Microbiol Mol Biol Rev. 2015;79(3):263–280. doi:10.1128/mmbr.00010-15.
- Lee S-J, Kelley BS, Damasceno CMB, John BS, Kim B-S, Kim B-D, Rose JKC. A functional screen to characterize the secretomes of eukaryotic pathogens and their hosts in planta. Mol Plant Microbe Interact. 2006;19(12):1368–1377. doi:10.1094/mpmi-19-1368.
- Jacobs KA, Collins-Racie LA, Colbert M, Duckett M, Golden-Fleet M, Kelleher K, Kriz R, LaVallie ER, Merberg D, Spaulding V, et al. A genetic selection for isolating cDNAs encoding secreted proteins. Gene. 1997;198(1–2):289–296. doi:10.1016/s0378-1119(97)00330-2.
- Lee S-J, Rose JKC. A yeast secretion trap assay for identification of secreted proteins from eukaryotic phytopathogens and their plant hosts. Methods in Mol Biol. 2006;19(12):1368–1377. doi:10.1094/mpmi-19-1368.
