ABSTRACT
Iron (Fe) is an important micronutrient for plant growth and development but any excess of Fe is toxic because of the Fe-dependent generation of reactive oxygen species (ROS). Thus, Fe homeostasis must be tightly regulated. In Arabidopsis thaliana, a cascade of transcription factors has been identified as involved in the regulation of this process by modulating the expression of genes related to Fe uptake, transport, and storage. Recently, it was demonstrated that in response to Fe deficiency, bHLH121/URI (UPSTREAM REGULATOR OF IRT1) directly activates the expression of several genes involved in this regulatory network. It was also shown that bHLH121 interacts with ILR3 (bHLH105) and its homologs. Herein it is shown that bHLH121 is necessary for the expression of the main markers of the plant responses to Fe excess, the ferritin genes (i.e. FER1, FER3, and FER4). bHLH121 regulates ferritin genes expression by directly binding to their promoters, at the same locus than the ILR3-PYE repressive complex. Therefore, this study highlight that bHLH121, PYE, and ILR3 form a chain of antagonistic switches that regulate the expression of ferritin genes. The implication of this finding is discussed.
Iron (Fe) is an important micronutrient for plant growth and development, as it serves as cofactors for numerous enzymes involved in various cellular processes such as photosynthesis, respiration, or the synthesis of amino acids.Citation1–Citation3 However, an excess of Fe is deleterious for plants because of its capacity to generate ROS.Citation4 Thus, the level of Fe in plant cells must be tightly regulated to avoid both Fe deficiency and Fe excess.
Fe uptake is the first limiting step for the maintenance of Fe homeostasis in plants. Non-grass species have evolved a reduction base mechanism allowing taking up the Fe present in the soil in the form of Fe(III)-chelates ().Citation6–Citation8 Once into the plant, Fe is transported to the different organs to be assimilated in several metalloproteins or stored in different cell compartments. Part of the Fe is transiently stored into ferritins whose expression is strongly induced in response to Fe excess.Citation9,Citation10, In Arabidopsis thaliana, there are four ferritin genes; three are expressed in vegetative tissues (i.e. FER1, FER3, and FER4) and one in seeds (i.e. FER2).Citation9,Citation11
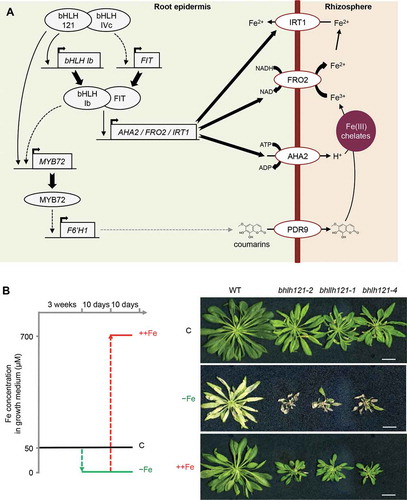
Figure 1. Loss-of-function of bHLH121 leads to severe iron deficiency symptoms that can be rescued by extra iron supply.
Maintaining Fe homeostasis necessitates the activity of several transcription factors (TFs) organized into an intricate regulatory network.Citation12,Citation13 In Arabidopsis, it involves 17 bHLH TFs, among which ILR3/bHLH105 (IAA-LEUCINE RESISTANT3) plays a dual role.Citation13 ILR3 physically interacts with its orthologs from the bHLH clade IVc (i.e. bHLH34, bHLH104, and bHLH115) to activate the Fe deficiency responses ().Citation14–Citation17 ILR3 also acts as a transcriptional repressor when it interacts with PYE/bHLH47 (POPEYE).Citation10 Among the ILR3-PYE targets are the ferritin genes (i.e. FER1, FER3, and FER4), the main markers of the plant response to Fe excess.Citation10
bHLH121 (URI, UPSTREAM REGULATOR OF IRT1) was identified as a novel TF that acts upstream the Fe homeostasis regulatory network, together with ILR3 and its orthologs, as a transcriptional activator.Citation15–Citation18 bhlh121 mutants display severe Fe deficiency symptoms, even in control Fe condition, that can be rescued by providing extra Fe supply ().Citation15–Citation17 In agreement with the upstream position of bHLH121 in the Fe homeostasis network, bhlh121 mutants are affected in all the aspects of the Fe-deficiency responses and the expression of several regulatory proteins and peptides involved in this network is impaired.Citation15–Citation17
Since bHLH121 controls Fe uptake, one may hypothesize that bHLH121 might directly or indirectly regulate the expression of ferritin genes. In order to validate this hypothesis, the expression of FER1, FER3, and FER4 was analyzed in wild type and three independent bhlh121 mutant lines by qRT-PCR.Citation16 For this purpose, seedlings were first grown for 7 days in both Fe sufficient (50 µM Fe) and Fe deficiency (0 µM Fe) conditions. Under Fe deficiency condition, expression analysis revealed that FER1 and FER4 mRNA accumulation was similar between the wild type and the mutants whereas FER3 mRNA accumulation was higher in the wild type than in the mutants (). These results suggest that bHLH121 has a positive effect on FER3 expression when Fe availability is low. In contrast, under Fe sufficient condition, the mRNA levels of the three ferritin genes were lower in the mutants than in the wild type. These results indicate that bHLH121 is necessary to maintain ferritin gene expressions when Fe availability is not limiting. These observations are in contrast with a previous study, based on the analysis of microarray data, showing that ferritin genes expression is not affected in bhlh121 mutants grown under Fe replete condition.Citation19 This apparent discrepancy might be explained by the higher sensitivity of the qRT-PCR method or to differences in the growth conditions.
Figure 2. bHLH121 is an activator of the Arabidopsis thaliana FER1, FER3 and FER4 expression.
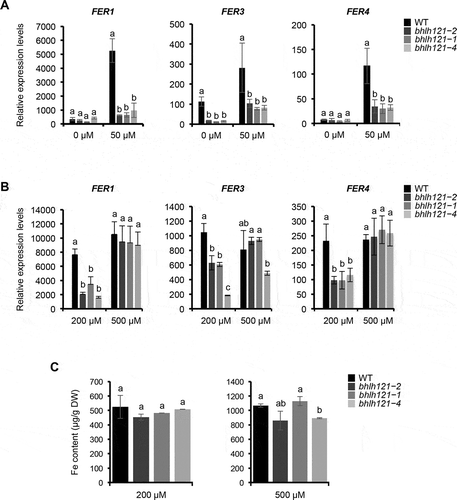
Considering the decrease in ferritins mRNA abundance observed in bhlh121 mutants when compared to the wild type (), it cannot be excluded that part of this diminution might be due to the reported differences in Fe accumulation between both genotypes (i.e. a reduction of about one-third in the mutant when compared to the wild type).Citation16 Therefore, similar experiments were conducted with seedlings grown under two different Fe excess conditions: 200 µM Fe (mid-Fe excess condition) and 500 µM Fe (high Fe excess condition). Under mid excess condition, the mRNA levels of the three ferritin genes were lower in the bhlh121 mutants than in the wild type () whereas Fe accumulation was similar between all genotypes (). This later result confirms that when Fe is not limiting bHLH121 is necessary for ferritin genes expression. Interestingly, under high Fe excess, the expression of ferritin genes was no longer different between the wild type and the bhlh121 mutants (). Fe accumulation was also similar between all genotypes (). In addition, at the rosette stage, no chlorosis was observed on the leaves of bhlh121 mutants submitted to Fe excess as it was previously reported for Arabidopsis plants deprived of ferritins (). Citation10 Taken together, these results indicate that bHLH121 is required to induce ferritin genes expression when Fe availability is in adequacy with the plant physiological needs. These data also suggest that under high Fe excess, ferritin genes expression relies on the IDRS (IRON-DEPENDENT REGULATORY SEQUENCE) signaling pathway.Citation19
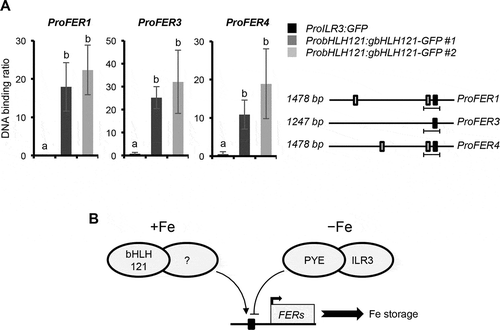
In order to confirm whether the regulation of ferritin genes expression by bHLH121 is direct or not, ChIP-qPCR experiments were conducted using two independent bhlh121 mutant lines complemented with the whole bHLH121 locus translationally fused to the GFP reporter gene (i.e. pbHLH121:gbHLH121-GFP in bhlh121-2).Citation16 This assay was centered on the promoter regions of the ferritin genes that contain the G-box motifs (CACGTG) known to be recognized by bHLH TFs. These loci were chosen for two reasons. First, because these G-boxes are directly targeted by the ILR3-PYE complex.Citation10 Second, because these G-boxes are located in nucleosome-free regions, suggesting the binding of regulatory proteins at the G-box loci.Citation10 ChIP-qPCR assays supported the in vivo binding of bHLH121 to the promoter of FER1, FER3, and FER4 (), confirming previous results obtained by ChIP-seq analysis.Citation19 ChIP-qPCR experiments also showed that bHLH121 binds to the promoter of ferritin genes at the same locus than the ILR3-PYE repressive complex.Citation10
Figure 3. bHLH121 is a direct activator of the Arabidopsis thaliana FER1, FER3 and FER4 expression.
While the transcript level and protein abundance of bHLH121 are not significantly affected by the Fe status, Fe availability affects the cellular localization of bHLH121 in Arabidopsis roots.Citation15,Citation16 Interestingly, the patterns of ILR3 and PYE accumulation in Arabidopsis root cells also depend on Fe availability. When Fe availability is not limiting, most bHLH121 accumulates in the stele, the site of ferritin gene expressions.Citation20 In this condition, ILR3 localizes in all root cells with higher abundance in the stele than in the epidermis and the cortex, whereas only traces of PYE are found in the stele.Citation5,Citation21 These observations indicate that, under Fe sufficient condition, the repressive activity of the ILR3-PYE complex on ferritin genes expression is low.Citation10,Citation22 It also supports that bHLH121 is a direct activator of ferritin genes expression. In contrast, when Fe availability is low, bHLH121 mainly localizes at the root epidermis and the cortex, the sites for Fe uptake, and ILR3 and PYE in all root cell types.Citation5,Citation16,Citation21 These observations are consistent with the repressive activity of the ILR3-PYE complex on ferritin genes expression and the positive role of bHLH121 on Fe uptake ().
The identification of the protein that interacts with bHLH121 to regulate ferritin genes expression is one of the main questions that remain to be solved (). It is unlikely that one of the clade IVc bHLH interact with bHLH121 to activate the expression of ferritin genes since ILR3 is a repressor of ferritin gene expressions and since the expression of ferritin genes is not lowered in the bhlh34, bhlh104, and bhlh115 loss-of-function mutants.Citation10 It is also unlikely that bHLH121 acts as homodimers since in vivo experiments suggest that bHLH121 cannot interact with itself.Citation16
Interestingly, the phosphorylation state of bHLH121 also depends on Fe availability.Citation15 Under Fe deficiency, the phosphorylated form of bHLH121 accumulates in roots. It is proposed that this mechanism allows the heterodimerization of bHLH121 with ILR3 and its three homologs, and thus the transcriptional activation of their target genes to activate the Fe deficiency responses.Citation15 Whether the phosphorylation of bHLH121 is necessary to activate the expression of ferritin genes will have to be demonstrated. Firstly, because bHLH121 activates the expression of ferritin genes in the stele when Fe availability is not limiting, and thus when bHLH121 phosphorylation is low. Second, because the phosphorylated form of bHLH121 is degraded when plants are recovering from Fe deficiency, a growth condition that strongly induces ferritin genes expression.Citation10,Citation15 The phosphorylation states of bHLH121 might rather be necessary to modify bHLH121 pattern of accumulation within the root cells in a Fe-dependent manner. In this hypothesis, which remains to be tested, there would be a balance between the phosphorylated bHLH121 form in the epidermis and the cortex and the non-phosphorylated form in the stele. The activity of FIT is also modulated by phosphorylation via the activity of CIPK11 (CBL-INTERACTING PROTEIN KINASE 11).Citation23,24 Whether or not CIPK11 or closely related kinases, might also phosphorylate bHLH121 and thus function as a coordinating factor for different aspects of Fe homeostasis is an appealing hypothesis that will have to be investigated.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed
Acknowledgments
FG is supported by a fellowship from the China Scholarship Council (CSC). KR was supported by fellowships from the ANR and the INRAE (department of “Biologie et Amélioration des Plantes”).
References
- Hänsch R, Mendel RR. Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr Opin Plant Biol. 2009;12:1–5. doi:10.1016/j.pbi.2009.05.006.
- Touraine B, Vignols F, Przybyla-Toscano J, Ischebeck T, Dhalleine T, Wu H-C, Magno C, Berger N, Couturier J, Dubos C, et al. Iron–sulfur protein NFU2 is required for branched-chain amino acid synthesis in Arabidopsis roots. J Exp Bot. 2019;70(6):1875–1889. doi:10.1093/jxb/erz050.
- Berger N, Vignols F, Przybyla-Toscano J, Roland M, Rofidal V, Touraine B, Zienkiewicz K, Couturier J , Feussner I, Santoni V, et al. Identification of client iron-sulfur proteins of the chloroplastic NFU2 transfer protein in Arabidopsis thaliana. J Exp Bot. 2020;71(14):4171–4187.doi: 10.1093/jxb/eraa166.
- Muckenthaler MU, Galy B, Hentze MW. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu Rev Nutr. 2008;28:197–213. doi:10.1146/annurev.nutr.28.061807.155521.
- Samira R, Li B, Kliebenstein D, Li C, Davis E, Gillikin JW, Long TA. The bHLH transcription factor ILR3 modulates multiple stress responses in Arabidopsis. Plant Mol Biol. 2018;97(4–5):297–309. doi:10.1007/s11103-018-0735-8.
- Tsai -H-H, Schmidt W. One way. Or another? Iron uptake in plants. New Phytol. 2017;214:500–505. doi:10.1111/nph.14477.
- Santi S, Schmidt W. Dissecting iron deficiency‐induced proton extrusion in Arabidopsis roots. New Phytol. 2009;183:1072–1084. doi:10.1111/j.1469-8137.2009.02908.x.
- Brumbarova T, Bauer P, Ivanov R. Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci. 2015;20:124–133. doi:10.1016/j.tplants.2014.11.004.
- Briat J-F, Duc C, Ravet K, Gaymard F. Ferritins and iron storage in plants. Biochimica Et Biophysica Acta (Bba)-gen Subj. 2010;1800:806–814. doi:10.1016/j.bbagen.2009.12.003.
- Tissot N, Robe K, Gao F, Grant‐Grant S, Boucherez J, Bellegarde F, Maghiaoui A, Marcelin R, Izquierdo E, Benhamed M, et al. Transcriptional integration of the responses to iron availability in Arabidopsis by the bHLH factor ILR3. New Phytol. 2019;223(3):1433–1446. doi:10.1111/nph.15753.
- Ravet K, Touraine B, Kim SA, Cellier F, Thomine S, Guerinot ML, Briat J-F, Gaymard F. Post-translational regulation of AtFER2 ferritin in response to intracellular iron trafficking during fruit development in Arabidopsis. Mol Plant. 2009;2:1095–1106. doi:10.1093/mp/ssp041.
- Connorton JM, Balk J, Rodríguez-Celma J. Iron homeostasis in plants–a brief overview. Metallomics. 2017;9:813–823. doi:10.1039/C7MT00136C.
- Gao F, Robe K, Gaymard F, Izquierdo E, Dubos C. The transcriptional control of iron homeostasis in plants: a tale of bHLH transcription factors? Front Plant Sci. 2019;10:6. doi:10.3389/fpls.2019.00006.
- Zhang J, Liu B, Li M, Feng D, Jin H, Wang P, Liu J, Xiong F, Wang J, Wang H-B, et al. The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell. 2015;27(3):787–805. doi:10.1105/tpc.114.132704.
- Kim SA, LaCroix IS, Gerber SA, Guerinot ML. The iron deficiency response in Arabidopsis thaliana requires the phosphorylated transcription factor URI. Proc National Acad Sci. 2019;116:24933–24942. doi:10.1073/pnas.1916892116.
- Gao F, Robe K, Bettembourg M, Navarro N, Rofidal V, Santoni V, Gaymard F, Vignols F, Roschzttardtz H, Izquierdo E, et al. The transcription factor bHLH121 interacts with bHLH105 (ILR3) and its closest homologs to regulate iron homeostasis in Arabidopsis. Plant Cell. 2020;32(2):508–524. doi:10.1105/tpc.19.00541.
- Lei R, Li Y, Cai Y, Li C, Pu M, Lu C, Yang Y, Liang G. bHLH121 functions as a direct link that facilitates the activation of FIT by bHLH IVc transcription factors for maintaining Fe homeostasis in arabidopsis. Mol Plant. 2020;3:634–649. doi:10.1016/j.molp.2020.01.006.
- Lockhart J. Personal trainer: BHLH121 functions upstream of a transcriptional network of heavy lifters involved in balancing iron levels. Plant Cell. 2020;32:293. doi:10.1105/tpc.19.00918.
- Briat J-F, Ravet K, Arnaud N, Duc C, Boucherez J, Touraine B, Cellier F, Gaymard F. New insights into ferritin synthesis and function highlight a link between iron homeostasis and oxidative stress in plants. Ann Bot. 2010;105(5):811–822. doi:10.1093/aob/mcp128.
- Reyt G, Boudouf S, Boucherez J, Gaymard F, Briat J-F. Iron-and ferritin-dependent reactive oxygen species distribution: impact on Arabidopsis root system architecture. Mol Plant. 2015;8:439–453. doi:10.1016/j.molp.2014.11.014.
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell. 2010;22:2219–2236. doi:10.1105/tpc.110.074096.
- Rampey RA, Woodward AW, Hobbs BN, Tierney MP, Lahner B, Salt DE, Bartel B. An Arabidopsis basic helix-loop-helix leucine zipper protein modulates metal homeostasis and auxin conjugate responsiveness. Genetics. 2006;174(4):1841–1857. doi:10.1534/genetics.106.061044.
- Gratz R, Manishankar P, Ivanov R, Köster P, Mohr I, Trofimov K, Steinhorst L, Meiser J, Mai H-J, Drerup M, et al. CIPK11-dependent phosphorylation modulates FIT activity to promote Arabidopsis iron acquisition in response to calcium signaling. Dev Cell. 2019;48(5):726–40. e10. doi:10.1016/j.devcel.2019.01.006.
- Gratz R, Brumbarova T, Ivanov R, Trofimov K, Tünnermann L, Ochoa‐Fernandez R, Blomeier T, Meiser J, Weidtkamp-Peters S, Zurbriggen MD, et al. Phospho‐mutant activity assays provide evidence for alternative phospho‐regulation pathways of the transcription factor FER‐LIKE IRON DEFICIENCY‐INDUCED TRANSCRIPTION FACTOR. New Phytol. 2020;225:250–267. doi:10.1111/nph.16168.
