ABSTRACT
Among all the major environmental challenges, drought stress causes considerable damage to plant growth and agricultural productivity. Drought stress directly promotes the accumulation of abscisic acid (ABA) via the activation of genes that encode enzymes involved in ABA biosynthesis, which protect the plant against water-limiting conditions. At the same time, the expression of genes that encode ABA-hydroxylases that inactivate the newly synthesized ABA, is repressed by drought stress. These phenomena occur through epigenetic modifications via the reversible processes of histone acetylation and deacetylation, also known as chromatin remodeling, which is an important regulatory mechanism that responds to various environmental stresses. Recently, we had reported that the chromatin remodeling complex HDA9-PWR-ABI4 promotes the development of drought tolerance through the deacetylation of CYP707A1/2 genes that encode the major enzymes involved in ABA catabolism. Here, we discuss the role of HDA9 and PWR in regulating drought stress by modulating the acetylation status of the CYP707A genes.
Introduction
Environmental stresses often lead to a series of physiological, biochemical, and molecular changes that result in serious damage to plant growth and development and severely affect agricultural productivity,Citation1 drought stress being a major threat among them.Citation2 To protect themselves against drought stress, plants have developed a series of sophisticated signaling cascades that help them complete their life cycle in situations where water availability is very low. These signaling cascades regulate ion homeostasis for osmotic adjustment and stomatal closure to reduce water loss through active transpiration.Citation3–Citation7 Both the above-mentioned mechanisms are controlled by the drought-induced phytohormone abscisic acid (ABA).Citation4,Citation5 Drought-induced accumulation of ABA is the result of two independent processes, i.e., the activation of inactive ABA and newly synthesized ABA.Citation6,Citation8,Citation9 ABA accumulation leads to the activation of a complex signal transduction pathway regulated by the combined activities of ABA receptors (PYR/PYLs), co-receptors (PP2Cs), kinases (SnRK2s), transcription factors (TFs), and ion channels.Citation10,Citation11
Drought stress promotes the activation of genes that encode key ABA biosynthesis enzymes, such as NCED3, ABA1, ABA2, and ABA3.Citation8,Citation12 In contrast, genes that encode enzymes that promote ABA inactivation, including the members of the CYP707A family, are suppressed under conditions of dehydration.Citation8 ABI4, an APETALA2 domain-containing TF, plays a major role in repressing the CYP707A1 and CYP707A2 genes. However, the detailed mechanism underlying the modulation of their chromatin structure has not yet been explored in detail.Citation13–Citation15
The increase or decrease in gene activity in response to environmental stresses is often associated with epigenetic changes such as histone acetylation and deacetylation. Histone acetylation leads to gene activation and deacetylation results in gene repression, and these processes are catalyzed by histone acetyltransferases (HATs) and HDACs, respectively.Citation16
Recently we reported that HDA9, an RPD3/HDA1-type class1 histone deacetylase and PWR (powerdress), a homolog of human NcoR1, interact with ABA4, which results in the formation of a chromatin remodeling complex.Citation9,Citation17 Further analysis revealed that the HDA9-PWR-ABI4 complex epigenetically regulates the expression of the CYP707A1 and CYP707A2 genes through histone deacetylation.Citation9,Citation17
PWR, HDA9, and ABI4 work together to regulate drought stress
Previous reports specified that HDA9 and PWR work together in a complex and regulate several physiological processes, including plant growth and development, flowering time, and leaf size and morphology.Citation18–Citation20 However, the mechanism by which this complex regulates abiotic stress responses was not explored well. Recently, we reported that HDA9 and PWR loss-of-function mutants show potent ABA insensitivity, including reduced sensitivity to ABA-mediated inhibition of seed germination and root growth, impaired ABA-induced stomatal closure, and reduced expression of ABA-responsive genes.Citation9,Citation17 Additionally, pwr and hda9 mutants show hypersensitive phenotypes under conditions of drought stress; these exhibit lower transcript levels of stress-responsive genes, impaired stomatal movement, and continuous water loss, thereby demonstrating that the two proteins positively regulate drought stress response.Citation9,Citation17
In order to explore the involvement of PWR and HDA9 in the regulation of ABA and drought stress responses in detail, a protein-protein interaction screen was performed; this screen showed that PWR and HDA9 specifically interact with ABI4.Citation9,Citation17 ABI4 is an APETALA2 domain-containing TF, which plays a major role in repressing CYP707A genes that code for ABA-hydroxylase enzymes which promote ABA inactivation.Citation13–Citation15 ABI4 was initially isolated as a major positive regulator of the ABA signaling pathway. However, several other reports indicated that besides ABA, ABI4 also plays a major role in key physiological processes, such as phytohormone cross-talk, flowering, and salt stress signaling.Citation21 More importantly, ABI4 is a versatile TF, which could either activate or repress the transcription of its target genes.Citation21 ABI4 binds specifically to ABRE elements and promotes the expression of several genes in response to abiotic stresses.Citation22 In contrast, the genes involved in ABA catabolism such as CYP707A genes, ethylene biosynthesis such as ACS genes, and retrograde signaling such as AOX1a are repressed by ABI4.Citation21 Interestingly the transcript level of genes repressed by ABI4, such as CYP707As, ACS4, and AOX1a, was significantly higher in pwr and hda9 mutants.Citation9,Citation17 As the PWR-HDA9 complex was identified as a repressor that modulates the acetylation status of numerous genes, the levels of total acetyl histone H3 (AcH3) were analyzed in pwr and hda9 mutants. As expected, the level of acetylated histone 3 significantly increased in pwr and hda9 mutants under conditions of dehydration stress, indicating that PWR and HDA9 modulate the acetylation status of drought-related genes. When we analyzed the AcH3 level in the promoter region of the CYP707A genes, we found that the AcH3 level was significantly higher in pwr and hda9 mutants than that in wild type.Citation9,Citation17 These findings suggest that PWR and HDA9 together with ABI4 form a chromatin remodeling complex involved in drought-stress‒induced repression of the CYP707A genes.Citation9,Citation17
Summary
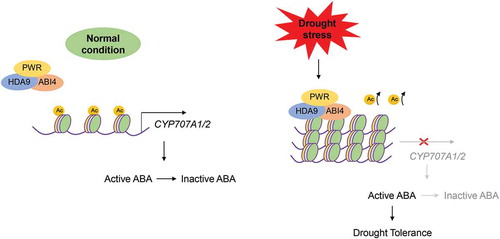
Apart from their role in regulating plant morphological and developmental processes, little is known about how HDA9 and PWR regulate signal transduction and chromatin modification in response to abiotic stresses. Based on recent findings, we propose a model, in which the CYP707A1 and CYP707A2 genes that encode (+)-abscisic acid 8′-hydroxylase under normal conditions are hyperacetylated and activated, thereby promoting ABA inactivation (). However, under conditions of drought stress, the PWR-HDA9-ABI4 complex binds to and promotes the deacetylation of the CYP707A1 and CYP707A2 genes in order to inhibit ABA-inactivation; thus the accumulation of active ABA promotes drought tolerance (). ABI4 has already been reported to repress CYP707A genes by directly binding to their promoters,Citation15 and inclusion of our results in this background suggests that ABI4 forms a complex with PWR and HDA9 and associates with the CYP707A1 and CYP707A2 promoters, resulting in histone deacetylation and inhibition of these genes.Citation9,Citation17 In conclusion, the HDA9-PWR-ABI4 complex plays a central role in the regulation of drought stress through the inhibition of ABA catabolism in order to promote the accumulation of active ABA, thereby protecting plants against dehydration. Further studies are required to understand how this complex modulates genes activated by ABI4 at a chromatin level.
Figure 1. Hypothetical model.
Author contributions
D-J.Y and A.A designed the study, performed the literature search, and wrote the manuscript.
Disclosure of potential conflict of interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the Next Generation Bio-Green21 Program, Rural Development Administration (RDA), Republic of Korea (SSAC, PJ01318201); the National Research Foundation of Korea (NRF) funded by the Korean Government (2019R1A2C2084096) and the Global Research Lab (2017K1A1A2013146).
Additional information
Funding
References
- Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167(2):1–3. doi:10.1016/j.cell.2016.08.029.
- Khan A, Pan X, Najeeb U, Tan DKY, Fahad S, Zahoor R, Luo H. Coping with drought: stress and adaptive mechanisms, and management through cultural and molecular alternatives in cotton as vital constituents for plant stress resilience and fitness. Biol Res. 2018;51:47. doi:10.1186/s40659-018-0198-z.
- Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA. 2009;106:8380–8385. doi:10.1073/pnas.0903144106.
- Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran LSP. ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 2014;202(1):35–49. doi:10.1111/nph.12613.
- Munemasa S, Hauser F, Park J, Waadt R, Brandt B, Schroeder JI. Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr Opin Plant Biol. 2015;28:154–162. doi:10.1016/j.pbi.2015.10.010.
- Kuromori T, Seo M, Shinozaki K. ABA transport and plant water stress responses. Trends Plant Sci. 2018;23:513–522. doi:10.1016/j.tplants.2018.04.001.
- Ali A, Kim J, Jan M, Khan HA, Khan IU, Shen M, Park J, Lim CJ, Hussain S, Baek D. Rheostatic control of ABA signaling through HOS15-mediated OST1 degradation. Mol Plant. 2019;12:1447–1462. doi:10.1016/j.molp.2019.08.005.
- Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Ann Rev Plant Biol. 2005;56(1):165–185. doi:10.1146/annurev.arplant.56.032604.144046.
- Baek D, Shin G, Kim MC, Shen M, Lee SY, Yun D-J. Histone deacetylase HDA9 with ABI4 contributes to abscisic acid homeostasis in drought stress response. Front Plant Sci. 2020;11:143. doi:10.3389/fpls.2020.00143.
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi:10.1038/nature08599.
- Ali A, Pardo JM, Yun D-J. Desensitization of ABA-signaling: the swing from activation to degradation. Front Plant Sci. 2020;11:379. doi:10.3389/fpls.2020.00379.
- Xiong L, Zhu J-K. Regulation of abscisic acid biosynthesis. Plant Physiol. 2003;133:29–36. doi:10.1042/bse0580029.
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell. 1998;10:1043–1054. doi:10.1105/tpc.10.6.1043.
- Okamoto M, Kushiro T, Jikumaru Y, Abrams SR, Kamiya Y, Seki M, Nambara E. ABA 9ʹ-hydroxylation is catalyzed by CYP707A in Arabidopsis. Phytochemistry. 2011;72:717–722. doi:10.1016/j.phytochem.2011.02.004.
- Shu K, Zhang H, Wang S, Chen M, Wu Y, Tang S, Liu C, Feng Y, Cao X, Xie Q. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet. 2013;9:e1003577. doi:10.1371/journal.pgen.1003577.
- Shahbazian MD, Grunstein M. Functions of site‐specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi:10.1146/annurev.biochem.76.052705.162114.
- Khan IU, Ali A, Khan HA, Baek D, Park J, Lim CJ, Zareen S, Jan M, Lee SY, Pardo JM. PWR/HDA9/ABI4 complex epigenetically regulates ABA dependent drought stress tolerance in arabidopsis. Front Plant Sci. 2020;11:623. doi:10.3389/fpls.2020.00623.
- Chen X, Lu L, Mayer KS, Scalf M, Qian S, Lomax A, Smith LM, Zhong X. POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in Arabidopsis. Elife. 2016;5:e17214. doi:10.7554/eLife.17214.
- Kim YJ, Wang R, Ga L, Li D, Xu C, Mang H, Jeon J, Chen X, Zhong X, Kwak JM. POWERDRESS and HDA9 interact and promote histone H3 deacetylation at specific genomic sites in Arabidopsis. Proc Natl Acad Sci USA. 2016;113:14858–14863. doi:10.1073/pnas.1618618114.
- Suzuki M, Shinozuka N, Hirakata T, Nakata MT, Demura T, Tsukaya H, Horiguchi G. OLIGOCELLULA1/HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES15 promotes cell proliferation with HISTONE DEACETYLASE9 and POWERDRESS during leaf development in Arabidopsis thaliana. Front Plant Sci. 2018;9:580. doi:10.3389/fpls.2018.00580.
- Chandrasekaran U, Luo X, Zhou W, Shu K. Multifaceted signaling networks mediated by abscisic acid insensitive 4. Plant Comm. 2020;1:100040. doi:10.1016/j.xplc.2020.100040.
- Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta. 2012;1819:86–96. doi:10.1016/j.bbagrm.2011.08.004.
