ABSTRACT
The gaseous phytohormone ethylene influences many aspects of plant life, including germination, fruit ripening, senescence, and stress responses. These diverse roles of ethylene occur in part through crosstalk with other phytohormones, which affects ethylene biosynthesis and signaling pathways. We have recently shown that the phytohormones, including gibberellic acid, abscisic acid, auxin, methyl jasmonate, and salicylic acid, regulate the stability of ACC synthases (ACSs), the rate-limiting enzymes in ethylene biosynthesis. Here, we report that treatment of etiolated Arabidopsis seedlings with strigolactone (SL) increases ethylene biosynthesis. SL does not influence ACS stability or ACS gene expression, but it increases the transcript levels of a subset of ACC oxidase (ACO) genes, thereby enhancing ethylene biosynthesis. Taken together with the results of our previous study, these findings demonstrate that most phytohormones differentially regulate ethylene biosynthesis in dark-grown Arabidopsis seedlings by affecting ACS stability and/or the transcript levels of ethylene biosynthesis genes.
The gaseous plant hormone ethylene influences a diverse array of plant growth, development, and biotic or abiotic stress responses through its crosstalk with other phytohormones and signaling pathways.Citation5–7 ACC synthases (ACS), the rate-limiting enzymes in ethylene biosynthesis, are central regulators that control how much ethylene is produced in plants by serving as convergence points for integrating other stimuli into the regulation of ethylene biosynthesis.Citation1,Citation5,Citation8
Ethylene is derived from the amino acid methionine via two intermediates, S-adenyl-L-methionine (SAM) and 1-aminocyclopropane-1-carboxylic acid (ACC).Citation9 SAM synthetases convert methionine to SAM, which is subsequently converted to ACC by ACC synthase (ACS). ACC is then oxidized to ethylene by ACC oxidase (ACO), a member of the oxygenase/oxidase superfamily of enzymes.Citation10 Most higher plant species contain a family of multiple ACS and ACO genes. For example, the Arabidopsis genome possesses five ACO and 12 ACS genes.Citation11,Citation12 Among 12 ACS genes, only eight encode functional ACS proteins which can be grouped into three different types (type-1, type-2, and type-3) based on the phosphorylation site in the C-terminal domain.Citation11,Citation13
Molecular genetics and biochemical studies have demonstrated that phosphorylation and dephosphorylation play a key role in regulating ACS stability. MITOGEN-ACTIVATED PROTEIN KINASE (MAPK)-mediated phosphorylation at the C-terminal domain of type-1 ACS increases the stability of the type-1 ACS, thereby increasing ethylene biosynthesis upon pathogen invasion.Citation14 ABA Insensitive 1 (ABI1), a protein phosphatase 2 C previously known to function as a negative regulator in ABA signaling, destabilizes type-1 ACS6 by antagonizing MAPK6 activity and by dephosphorylating the C-terminal domain of ACS6.Citation15 Furthermore, the degradation of type-3 ACS7 is hampered in PP2C type A knockout plants, suggesting the role of PP2Cs in ACS stability regulation.Citation16 Similarly, a protein phosphatase 2A ROOTS CURL IN 1-N-NAPHTHYLPHTHALAMIC ACID 1 (RCN1), promotes the degradation of a subset of ACS proteins.Citation17 By contrast to the positive role of phosphorylation on ACS stability, CASEIN KINASE 1.8-mediated phosphorylation of ACS5, ACS9, and ACS6 promotes the degradation of these ACS isoforms.Citation18 Compared to ACS, less is known about the role of post-translational modification on ACO stability. However, recent studies have shown that the activity of ACO is also regulated by post-translational modification.Citation19,Citation20 Sulfhydration is one such regulation that decreases the activity of tomato ACO1 and ACO2 via the sulfhydration of cysteine residues of the ACO isoforms, demonstrating the role of post-translational control on ACO in plants.Citation21
In addition to phosphorylation, the degradation of ACS via the ubiquitin-proteasome pathway is another common regulatory mechanism that controls ethylene biosynthesis. ETHYLENE-OVERPRODUCING 1 (ETO1) and its two paralogs ETO1-LIKE 1 (EOL1) and EOL2 specifically target type-2 ACS for degradation via the 26S proteasome pathway.Citation22,Citation23 ETO1 contains a BTB/TRP motif and serves as an adaptor protein for the CULLIN3 (CUL3) E3 ligase complex and imparts substrate specificity to CUL3.Citation13 Type-1 ACS stability is also regulated by the 26S proteasome, but the cognate E3 ligases have not yet been identified. Unlike the type-1 and type-2 ACSs, type-3 ACS does not contain any known regulatory motifs or phosphorylation sites. However, the stability of ACS7 is regulated by a RING-type E3 ligase XBAT32 and ABI1, a protein phosphatase 2 C, demonstrating that type-3 ACS stability is also under the control of phosphorylation and ubiquitination.Citation16,Citation24
Ethylene biosynthesis appears to be regulated by many external and internal stimuli. However, surprisingly, only a handful of stimuli have been identified and characterized that govern the stability of ACS.Citation1,Citation4 Phytohormone is one of the stimuli that influence the stability of ACS. For example, cytokinins and brassinosteroids (BRs) have long been known as positive regulators of ACS stability.Citation1,Citation4 We have also previously demonstrated a similar regulation of ACS stability by other phytohormones, including gibberellic acid (GA), abscisic acid (ABA), auxin, methyl jasmonate (MJ), and salicylic acid (SA), with distinct effects on different ACS isoforms.Citation1 For instance, SA increases the stability of type-2 ACS, whereas it promotes the degradation of type-1 ACS. MJ and ABA increase type-2 ACS stability, but neither hormone has any effect on the stability of type-1 ACS. Furthermore, none of these phytohormones influences type-3 ACS stability. It is worth noting that the insensitivity of the stability of the type-3 ACS7 isoform to most phytohormones may be related to the nature of the N-terminus-tagged fusion of ACS7 protein used in the study, which blocks the degradation sequence located in the N-terminal domain.Citation25 Further studies on the role of phytohormones on the C-terminal fusion of ACS7 will provide more insights into the phytohormone-mediated regulation of ACS stability.
Strigolactones (SLs), a group of carotenoid-derived phytohormones, were initially characterized as root-derived compounds that stimulate the germination of the seeds of parasitic weeds such as Striga, Orobanche, and Phelipanche species.Citation26–28 SLs also play a pivotal role in plant growth and developmental processes, including the determination of root and shoot architectures, flower development, and leaf senescence.Citation28 Many of these processes are associated with crosstalk between SLs and other phytohormones, including ethylene. For example, leaf senescence is strongly accelerated by the combination of SL with ethylene, not by SL alone, suggesting that SL stimulates leaf senescence through the action of ethylene. Conversely, ethylene treatment enhances SL biosynthesis by increasing expression of the strigolactone biosynthesis genes MORE AXIALLY GROWTH3 (MAX3) and MAX4 during leaf senescence.Citation29 SL and ethylene also regulate root hair elongation. The study showed that ethylene is epistatic to SL and ethylene biosynthesis is required for SL-mediated root hair elongation.Citation30 Roots treated with the ethylene biosynthesis inhibitor AVG do not elongate even in the presence of GR24, a synthetic SL analog, while GR24 alone increases root hair elongation. These results indicate that ethylene plays a positive role in SL biosynthesis. However, a possible role of SL on the biosynthesis of ethylene remains to be established.
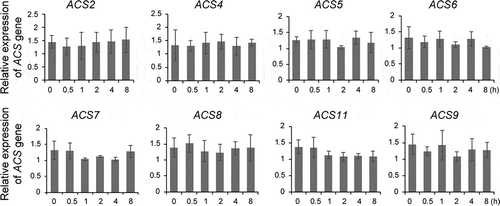
We did not examine the role of SL as regulators of the stability of ACS in dark-grown seedlings in our previous study.Citation1 In the present study, we expanded our analysis by examining the effect of SL on the stability of ACS using dark-grown Arabidopsis seedlings expressing Myc-tagged ACS proteins. Similar to several other phytohormones in the previous study, treatment of GR24 increased ethylene biosynthesis in dark-grown wild-type and seedlings expressing Myc-ACS2 (type-1), Myc-ACS5 (type-2), or MycACS7 (type-3). The levels of ethylene in wildtype and Myc-ACS overexpression lines increased in proportion to the increased concentration of GR24 (). We also examined whether the GR24-mediated increase in ethylene biosynthesis is due to an increase in the abundance of ACS proteins by determining the steady-state levels of ACS. An increased concentration of GR24 did not affect the steady-state levels of all three types of ACS proteins (). Furthermore, GR24 did not affect ACS gene expression, regardless of treatment duration (). These results indicated that ACS is not the target of GR24 in the ethylene biosynthesis pathway, although it is possible that GR24 may control the activity of ACS activity via post-translational modification. Furthermore, the involvement of other ACS isoforms that have not been examined in this study may attribute to the GR24-mediated increase in ethylene production. Further study will answer these questions. In contrast to our result, a previous study showed that SL enhances ACS2 gene expression in Arabidopsis roots.Citation30 This discrepancy may result from differences in the experimental conditions, including the growth conditions (light-grown vs dark-grown), the concentration of SL, and the treatment duration.
Figure 1. GR24 enhances ethylene biosynthesis in dark-grown Arabidopsis seedlings but does not influence ACS stability. (a) Seedlings were grown in capped vials containing growth media with the indicated concentration of GR24 for 3 d and the accumulated ethylene was measured by gas chromatography. Error bars indicate SD; n = 3. (b) Seedlings expressing myc-ACS2, myc-ACS5, or myc-ACS7 were grown on growth media with the indicated concentrations GR24 for 3 d. Total protein extracts from the seedlings were analyzed by immunoblotting using an anti-Myc or an anti-HSC70 antibody as a loading control. The Myc-ACS bands were normalized to the HSC70 control, and these values were then normalized to the “no GR24” control value, which was set to 1. The numbers below the gel panels indicate the relative ratios of the band intensities. * P < .05, Student’s t-test
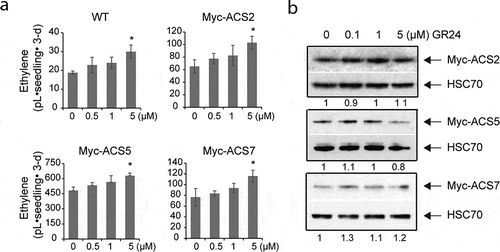
Figure 2. GR24 does not influence ACS gene expression. Quantitative RT-PCR analysis of ACS genes in wild-type seedlings treated with 5 μM GR24. Seedlings were harvested at the indicated times after GR24 treatment and used for analysis. Error bars indicate the standard deviation (SD) for three biological replicates
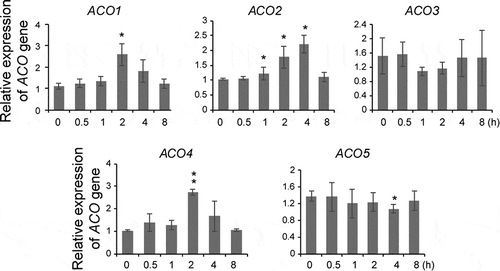
We additionally investigated whether ACO might be a target of SL as increased ACO transcript levels or ACS stability also leads to an increase in ethylene biosynthesis. Unlike its effect on the ACS genes, GR24 increased the transcript levels of a subset of ACO genes (ACO1, ACO2, and ACO4) approximately 2–4 fold within 2 to 4 h after the treatment (). By contrast, no changes were observed in the transcript levels of the other ACO genes (ACO3 and ACO5) although a slight decrease in ACO5 transcript was observed at 4 h after the treatment (). These results demonstrated that the GR24-mediated increase in ethylene biosynthesis was a result of an increase in ACO gene expression, although the possibility remains for GR24-mediated stabilization of ACO proteins.
The observation of downregulation of ACO transcripts, with no effect on ACS stability, by SL was reminiscent of the effects of MJ and ABA, although these hormones downregulate ACO transcripts.Citation1 However, unlike MJ and ABA, which showed different effects on the stability of type-1 and type-2 ACS, SL did not influence the stability of any of the three ACS types in dark-grown seedlings. Taken together, these results demonstrated that SL regulates ethylene biosynthesis by enhancing the transcript levels of the ACO genes without influencing ACS stability. Elucidation of the effects of phytohormones on ACS stability in light-grown seedlings or in response to stress conditions will provide more insight into the differential role of most phytohormones on the regulation of ethylene biosynthesis.
Figure 3. GR24 differentially regulates ACO gene expression in dark-grown seedlings. Quantitative RT-PCR analysis of ACO genes in wild-type seedlings treated with 5 μM GR24. Seedlings were harvested at the indicated times after GR24 treatment and used for analysis. Error bars represent the standard deviation (SD) for three replicates. * P < .05, ** P < .005, Student’s t-test
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana Col-0 was used as the wildtype (WT) reference throughout the study. All plants were grown in either long-day or short-day conditions at 22ºC ± 2ºC or in vitro on Murashige and Skoog (MS) Basal Medium supplemented with 0.8% plant agar (pH 5.7) in a continuous light chamber at 21°C. Transgenic Arabidopsis lines used were previously generated as 35S CaMV constitutive promoter-driven dexamethasone (Dex)-inducible Myc-tagged ACS2, ACS5, or ACS7.Citation1 Briefly, the coding region of each ACS gene was cloned into the Gateway pENTR vector and transferred to a modified Gateway‐compatible version of the binary GVG vector pTA7002Citation2 to which a 6‐myc cassette was added. Col‐0 plants were transformed by the floral dip methodCitation3 and transformants were selected on MS medium containing hygromycin.
Treatment of seedlings with GR24 for protein analysis and RT-PCR
For protein analysis, seedlings expressing Myc-ACS2, Myc-ACS5, or Myc-ACS7 were grown on growth medium containing 20, 40, or 5 nM Dex in the absence or presence of GR24 (PhytoTechnology Laboratories) for 3 d in the dark. For quantitative RT-PCR analysis, 3-day-old dark-grown wild type seedlings were treated with 5 μM GR24 for the indicated times and harvested for analysis.
Protein analysis
Proteins were extracted from 3‐day‐old etiolated seedlings in 2× SDS buffer (65.8 mM Tris HCl, pH 6.8, 26.3% Glycerol (w/v), 2.1 % SDS, 0.01% Bromophenol blue) and ground with a mini‐pestle. Total protein extracts were incubated at 95°C for 3 min and then centrifuged for 3 min at room temperature at 16 000 g in a microcentrifuge. Immunoanalysis was performed using an anti-Myc antibody (Sigma-Aldrich) and HSC70 (Enzo) as a loading control.
RNA extraction and quantitative RT-PCR
Total RNA was prepared using the RNeasy Plant Mini Kit (QIAGEN) and reverse transcribed using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturers’ instructions. Quantitative RT-PCR was performed using PowerUPTMSYBRGreen Master Mix (Applied Biosystems). The primers used are listed in Table S1. Three biological replicates were analyzed with three technical replicates per sample. The relative expression of the candidate genes was normalized to β-tubulin.
Measurements of ethylene production
Ethylene measurements were performed as previously described.Citation4 Surface-sterilized seeds were germinated in 22 mL gas chromatography vials containing 3 mL of a medium consisting of half-strength MS medium/1% sucrose/0.8% BactoTM agar, Dex, and GR24. After 3 d of stratification at 4°C in darkness, the vials containing Arabidopsis seeds were exposed to light for 3 h, capped, incubated in dark for 3 d and the accumulated ethylene was measured by gas chromatography using a Shimadzu GC2010 Plus capillary gas chromatography system with an HS-20 headspace autosampler. Ethylene concentration was calculated in pL•seedling•3 days. All genotypes and treatments were measured from three biological replicates and an average and SD were calculated.
Author contributon
GMY conceived the experiments; HYL conducted most experiments; GMY and HYL wrote the manuscript.
Data availability
All data discussed in the paper are available in the main text and Supplementary Information.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supporting information
Supplemental Table 1. The list of primers used in this study.
Supplemental Material
Download PDF (67.8 KB)Acknowledgments
This work was supported by the start-up fund from Purdue University and a grant from NSF (MCB-1817286) to GMY.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
Additional information
Funding
References
- Lee HY, Chen YC, Kieber JJ, Yoon GM. Regulation of the turnover of ACC synthases by phytohormones and heterodimerization in Arabidopsis. Plant J. 2017;91(3):1–5. doi:https://doi.org/10.1111/tpj.13585.
- Aoyama T, Chua NH. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11(3):605–612. doi:https://doi.org/10.1046/j.1365-313X.1997.11030605.x.
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi:https://doi.org/10.1046/j.1365-313x.1998.00343.x.
- Hansen M, Chae HS, Kieber JJ. Regulation of ACS protein stability by cytokinin and brassinosteroid. Plant J. 2009;57(4):606–614. doi:https://doi.org/10.1111/j.1365-313X.2008.03711.x.
- Abeles FB. Ethylene in plant biology. Vol. 302, New York, NY: Academic; 1973.
- Ku YS, Sintaha M, Cheung MY, Lam HM. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int J Mol Sci. 2018;19(10):3206. doi:https://doi.org/10.3390/ijms19103206.
- Verma V, Ravindran P, Kumar PP. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016;16:86. doi:https://doi.org/10.1186/s12870-016-0771-y.
- Seo DH, Yoon GM. Light-induced stabilization of ACS contributes to hypocotyl elongation during the dark-to-light transition in Arabidopsis seedlings. Plant J. 2019;98(5):898–911. doi:https://doi.org/10.1111/tpj.14289.
- Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Ann Rev Plant Physiol. 1984;35:155–189. doi:https://doi.org/10.1146/annurev.pp.35.060184.001103.
- Bidonde S, Ferrer MA, Zegzouti H, Ramassamy S, Latche A, Pech JC, Hamilton AJ, Grierson D, Bouzayen M. Expression and characterization of three tomato 1-aminocyclopropane-1-carboxylate oxidase cDNAs in yeast. Eur J Biochem. 1998;253(1):20–26. doi:https://doi.org/10.1046/j.1432-1327.1998.2530020.x.
- Houben M, Van de Poel B. 1-Aminocyclopropane-1-Carboxylic Acid Oxidase (ACO): the Enzyme that makes the plant hormone ethylene. Front Plant Sci. 2019;10:695. doi:https://doi.org/10.3389/fpls.2019.00695.
- Vanderstraeten L, Van Der Straeten D. Accumulation and transport of 1-Aminocyclopropane-1-Carboxylic Acid (ACC) in plants: current status, considerations for future research and agronomic applications. Front Plant Sci. 2017;8:38. doi:https://doi.org/10.3389/fpls.2017.00038.
- Chae HS, Kieber JJ. Eto brute? Role of ACS turnover in regulating ethylene biosynthesis. Trends Plant Sci. 2005;10(6):291–296. doi:https://doi.org/10.1016/j.tplants.2005.04.006.
- Han L, Li GJ, Yang KY, Mao G, Wang R, Liu Y, Zhang S. Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J. 2010;64(1):114–127. doi:https://doi.org/10.1111/j.1365-313X.2010.04318.x.
- Ludwików A, Cieśla A, Kasprowicz-Maluśki A, Mituła F, Tajdel M, Gałgański Ł, Ziółkowski PA, Kubiak P, Małecka A, Piechalak A, et al. Arabidopsis protein phosphatase 2C ABI1 interacts with type I ACC synthases and is involved in the regulation of ozone-induced ethylene biosynthesis. Mol Plant. 2014;7(6):960–976. doi:https://doi.org/10.1093/mp/ssu025.
- Marczak M, Cieśla A, Janicki M, Kasprowicz-Maluśki A, Kubiak P, Ludwików A. Protein phosphatases type 2C Group A interact with and regulate the stability of ACC Synthase 7 in Arabidopsis. Cells. 2020;9(4):978. doi:https://doi.org/10.3390/cells9040978.
- Skottke KR, Yoon GM, Kieber JJ, DeLong A. Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet. 2011;7(4):e1001370. doi:https://doi.org/10.1371/journal.pgen.1001370.
- Tan ST, Xue HW. Casein kinase 1 regulates ethylene synthesis by phosphorylating and promoting the turnover of ACS5. Cell Rep. 2014;9:1692–1702. doi:https://doi.org/10.1016/j.celrep.2014.10.047.
- Dixon DP, Skipsey M, Grundy NM, Edwards R. Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol. 2005;138(4):2233–2244. doi:https://doi.org/10.1104/pp.104.058917.
- Friso G, van Wijk KJ. Posttranslational protein modifications in plant metabolism. Plant Physiol. 2015;169(3):1469–1487. doi:https://doi.org/10.1104/pp.15.01378.
- Jia H, Chen S, Liu D, Liesche J, Shi C, Wang J, Ren M, Wang X, Yang J, Shi W, et al. Ethylene-induced hydrogen sulfide negatively regulates ethylene biosynthesis by persulfidation of ACO in tomato under osmotic stress. Front Plant Sci. 2018;9:1517. doi:https://doi.org/10.3389/fpls.2018.01517.
- Yoshida H, Nagata M, Saito K, Wang KL, Ecker JR. Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol. 2005;5:14. doi:https://doi.org/10.1186/1471-2229-5-14.
- Wang KL, Yoshida H, Lurin C, Ecker JR. Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature. 2004;428(6986):945–950. doi:https://doi.org/10.1038/nature02516.
- Lyzenga WJ, Booth JK, Stone SL. The Arabidopsis RING-type E3 ligase XBAT32 mediates the proteasomal degradation of the ethylene biosynthetic enzyme, 1-aminocyclopropane-1-carboxylate synthase 7. Plant J. 2012;71:23–34.
- Xiong L, Xiao D, Xu X, Guo Z, Wang NN. The non-catalytic N-terminal domain of ACS7 is involved in the post-translational regulation of this gene in Arabidopsis. J Exp Bot. 2014;65(15):4397–4408. doi:https://doi.org/10.1093/jxb/eru211.
- Humphrey AJ, Beale MH. Strigol: biogenesis and physiological activity. Phytochemistry. 2006;67(7):636–640. doi:https://doi.org/10.1016/j.phytochem.2005.12.026.
- Burger M, Chory J. The many models of strigolactone signaling. Trends Plant Sci. 2020;25(4):395–405. doi:https://doi.org/10.1016/j.tplants.2019.12.009.
- Al-Babili S, Bouwmeester HJ. Strigolactones, a novel carotenoid-derived plant hormone. Annu Rev Plant Biol. 2015;66(1):161–186. doi:https://doi.org/10.1146/annurev-arplant-043014-114759.
- Ueda H, Kusaba M. Strigolactone regulates leaf senescence in concert with ethylene in arabidopsis. Plant Physiol. 2015;169(1):138–147. doi:https://doi.org/10.1104/pp.15.00325.
- Kapulnik Y, Resnick N, Mayzlish-Gati E, Kaplan Y, Wininger S, Hershenhorn J, Koltai H. Strigolactones interact with ethylene and auxin in regulating root-hair elongation in Arabidopsis. J Exp Bot. 2011;62(8):2915–2924. doi:https://doi.org/10.1093/jxb/erq464.
