ABSTRACT
Plant potexvirus and potyvirus infection can trigger endoplasmic reticulum (ER) stress. ER stress signaling increases the expression of cytoprotective ER-chaperones, especially the BiP chaperones which contribute to pro-survival functions when plants are subjected to infection. The inositol requiring enzyme (IRE1) is one ER stress sensor that is activated to splice the bZIP60 mRNA which produces a truncated transcription factor that activates gene expression in the nucleus. The IRE1/bZIP60 pathway is associated with restricting potyvirus and potexvirus infection. Recent data also identified the IRE1-independent UPR pathways led by bZIP28 and bZIP17 contribute to potexvirus and potyvirus infection. These three bZIP pathways recognize cis-regulatory elements in the BiP promoters to enhance gene expression. BiP is part of a negative feedback loop that regulates the activities of the ER stress transducers IRE1, bZIP28, and bZIP17 to block their activation. We discuss a model in which bZIP60 and bZIP17 synergistically induce BiP and other genes restricting Plantago asiatica mosaic virus (PlAMV; a potexvirus) infection while bZIP60 and bZIP28 independently induce genes supporting PlAMV infection. Regarding Turnip mosiac virus (TuMV, a potyvirus) infection, bZIP60 and bZIP28 serve to repress local and systemic infection. Finally, tauroursodeoxycholic acid treatments were used to demonstrate that the protein folding capacity significantly influences PlAMV accumulation.
IRE1-dependent and independent UPR pathways
The proper regulation of transcription, protein folding and maturation are necessary for cells to perform a myriad of normal processes and respond to plant virus infection. Protein folding and maturation occurs in the endoplasmic reticulum (ER) and is carried out by protein chaperones of the quality control (ERQC) such as the ER lumen binding protein (BiP), calnexin, calreticulin, and protein disulfide isomerase (PDI). BiP is an important controller of protein folding efficiency by helping to rachet nascent proteins into the ER and chaperoning their proper folding in an ATP-dependent manner. These ER resident chaperones, including BiP also directs degradation of malformed proteins (known as ERAD) that cannot be refolded.Citation1–6 The ERQC machinery is necessary for plant growth and development, as well as various kinds of environmental acclimation. When plants are under environmental stress from biotic or abiotic stressors, there are adaptive responses to maintain the ERQC, and this is known as the unfolded protein response (UPR). The UPR has four major attributes: 1) provides protein quality control in the ER, 2) initiates a signaling cascade to upregulate genes needed for chaperoning protein folding, 3) regulates autophagy and programmed cell death (PCD), and 4) is shown in mammals but not plants to expands ER membranes to accommodate the increased demands for protein synthesis.Citation7–12
In plants and mammals, BiPs act as negative feedback regulators of the UPR (). The upregulation of BiP at the transcription level is triggered by the accumulation of malformed proteins in the ER. The primary mediator of this response is the inositol requiring enzyme (IRE1) which is a transmembrane kinase/endoribonuclease occurring in mammals, plants, and yeast. BiP binds to the ER lumen domain of IRE1 and prevents its activation.Citation13,Citation14 Yeast has one isoform of IRE1, while mammals and plants have two isoforms that are involved in UPR. Dissociation of BiP or binding of unfolded proteins to IRE1 allows for face-to-face dimerization of IRE1 and trans-autophosphorylate, thus activating its endoribonuclease activity. This activity splices the XBP1 mRNA in mammals and bZIP60 mRNA in plants to remove a 23–26 nt intron to produce a functioning transcription factor (XBP1s or bZIP60s). The XBP1s and bZIP60s transcription factors mobilize to the nucleus and induce gene expression through the UPRE/ERSE cis-element in specific gene promoters, including the BiP promoters.Citation15–19
Figure 1. UPR mediated suppression of plant virus infection. The IRE1a/IRE1b-dependent, bZIP28/bZIP17 dependent pathways are routes leading to transcriptional activation of genes that regulate pro-survival and pro-death events. BiP is an ER resident molecular chaperone that is a master regulator of these ER stress transducers to block their activation. BiP also functions in the ER lumen to facilitate protein folding in an ATPase dependent manner. The bZIP60, bZIP28, and bZIP17 bind to the BiP promoter and increasing its expression to enhance protein folding in the ER lumen and to serve as part of a negative feedback loop to block further activation of these stress transducers in the ER. IRE1a/IRE1b endonuclease activity splice the bZIP60 mRNA to produce a transcription factor that is mobilized to the nucleus. NAC089 and NAC103 are activated to regulate programmed cell death and enhance protein quality control. bZIP60 and bZIP17 form complexes that activate unknown genes that limit PlAMV infection. The bZIP60 and bZIP28 activate unknown genes that support PlAMV infection but limit TuMV infection. Overexpression of BiP or enhancing protein folding capacity through TUDCA limits virus infection and suppressed PCD
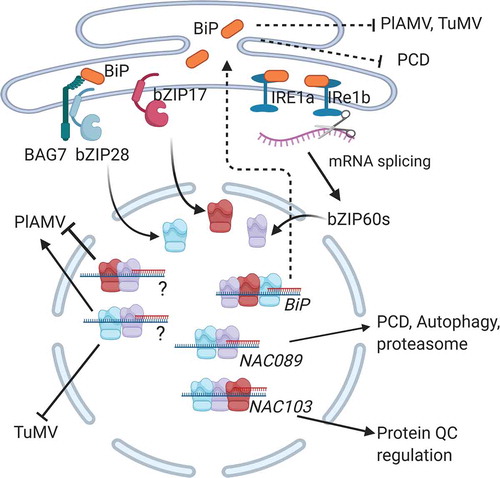
Additional layers of input for robust BiP expression are provided by IRE1-independent UPR pathways led by the bZIP28 and bZIP17 ER transmembrane proteins, which also activate BiP genes in plants.Citation19–21 Within the ER, bZIP28 associates with BiP and with the Bcl-2–associated athanogene 7 (BAG7) under regular conditions and dissociates upon ER stress ().Citation22 During unstressed conditions, BiP prevents the mobilization of bZIP28 and bZIP17 out of the ER.Citation23,Citation24 In response to ER stress, bZIP28 and bZIP17 are released from BiP and translocated to golgi via COPII vesicles followed by sequential cleavage of the cargo to the transmembrane domains by site-1 protease (S1P) and site-2 protease (S2P). This permits them to translocate into the nucleus (). The bZIP28 and bZIP17 transcription factors bind to UPRE or ERSE cis-regulatory elements to activate the expression of target genes including BiPs. The bZIP28, bZIP60, and bZIP17 can each bind to promoter elements but they can also combine with related bZIP factors such as bZIP49 or NF-Y factors, into heterodimeric transcription complexes that upregulate expression of ER stress-related genes.Citation25
UPR pathways are activated by viral proteins in the ER
Both in mammals and plants, RNA viruses have been shown to activate the IRE1-XBP1/bZIP60 pathway to cope with ER stress during infection. In mammals, members of the Flaviviridae family such as the Dengue virus (DENV), Japanese encephalitis virus (JEV), West Nile virus (WNV) and Zika virus (ZIKV) depend upon the ER for their translation, replication, and packaging.Citation26 These viruses also encode small hydropohic membrane-anchored proteins which associate with the ER and trigger the XBP1 signaling pathway.Citation26–29 Researchers argue that ER stress is caused by the ER-tropic nature of flaviviruses which disrupts the normal post-translational functions of the ER. The UPR is activated to attenuate the cytopathic effects of ER stress so that viruses have more time and space for replication to occur.Citation26 This is supported by many studies including an example for JEV showing that experimentally depleting XBP1 reduces the levels of the autophagy effectors ATG3 and BECLIN1, prevents autophagy, and enhances JEV-induced cell death.Citation30 Flaviviruses are also reported to activate the UPR to ensure proper protein folding and the degradation of malformed proteins in the ER.Citation26,Citation28 In plants, members of the genera Potexvirus, Potyvirus, and Figivirus are also ER-tropic, relying extensively on this membranous network for translation, replication, and cell-to-cell movement.Citation8,Citation27,Citation31–33 These viruses encode small hydrophobic membrane-anchored proteins that associate with the ER and trigger the -bZIP60 signaling pathway. Plant viral activation of the bZIP60 pathway leads to the expression of cellular chaperones in Nicotiana benthamiana and Arabidopsis (ecotype Col-0).Citation16,Citation34–36 Comparing these plant viruses and the mammalian flaviviruses, we speculate that that the UPR in plants protects cells against cytotoxic death and creates the space and time for virus replication to occur. In this regard, cells might recognize the small hydrophobic viral proteins along the ER as constitutively misfolded proteins causing UPR activation. However, further studies are needed to uncover how these viral proteins activate UPR in plants.
Studies also show that there may be a direct effect of the UPR on plant virus replication, cell-to-cell movement or systemic transport through the vasculature which requires further investigations to understand. Plants have two IRE1 genes known as IRE1a and IRE1b and recent investigations indicate that these factors differentially recognize the small hydrophobic membrane anchor proteins of potexviruses and potyviruses. Infectious clones of the potexvirus plantago asiatica mosaic virus containing the GFP gene (PlAMV-GFP) and a clone of the potyvirus turnip mosaic virus containing the GFP gene (TuMV-GFP) were was used to monitor virus infection in wild-type and mutant Arabidopsis plants. Immunoblots detecting the viral coat protein and GFP were also used to monitor virus levels in mutant Arabidopsis plants. PlAMV-GFP accumulation was higher in ire1a and bzip60 mutant, but not ire1b mutant Arabidopsis plants indicating that IRE1a-bZIP60 signaling normally suppresses PlAMV infection. For TuMV-GFP infection, GFP and coat protein levels were preferentially elevated in ire1b plants and were further enhanced in bzip60 and ire1a/ire1b plants, suggesting both IRE1 isoforms restrict TuMV infection.Citation35–38 We still do not understand why IRE1a is solely important for PlAMV infection while IRE1a and IRE1b play overlapping roles in responding to TuMV infection when both proteins can activate bZIP60. Further studies are needed to understand if IRE1 and bZIP60 restriction of either virus is due to a direct interaction between these cellular proteins and the viral replication or cell-to-cell movement machinery, or if the IRE1 endonuclease activity can attack viral transcripts, or whether signal transduction activates defense genes that suppress virus infection.
A recent study suggests that signal transduction activates genes that regulate virus infection is a plausible hypothesis. We recently reported that the IRE1-independent UPR pathways have some overlapping ability to favor or restricts plant virus infection.Citation36 PlAMV-GFP and TuMV-GFP were reported to induce expression of bZIP28 and bZIP17 alongside bZIP60 in wild-type Col-0 plants within 2 days of inoculation.Citation36 In genetic studies comparing the bzip60, bzip28, and bzip17 KO mutant Arabidopsis, PlAMV-GFP fluorescence reached higher levels in inoculated and systemic leaves in bzip60 and bzip17 mutants and was unaltered in bzip28 mutants. These data suggested a model in which bZIP60 and bZIP17 synergistically induce genes that restrict PlAMV-GFP infection while bZIP60 and bZIP28 induce separate genes that support infection (). On the other hand, TuMV-GFP infection was elevated in bzip60 and bzip28 KO plants and is unaltered in bzip17 KO plants. In this case, bZIP60 and bZIP28 combine to repress TuMV-GFP, while bZIP17 does not appear to play any contributing role (). These differential responses point to additional layers of regulation that help the cell to cope with virus-induced ER stress.
Until now research has demonstrated roles of the bZIP60-, bZIP28- and bZIP17- led pathways for regulating the expression of BiP genes and other ER-resident chaperones that are important for protein quality control and protein maturation during virus-induced ER stress. However, it is more challenging to identify additional genes that either act directly to affect viral infection or to create a cellular environment to cope with infection. In Gayral et al. (2020), we showed activation of the NAC103 and NAC089 transcription factors represent a second tier of even more complex genetic responses to plant viruses. NAC103 is responsive to bZIP60, but not bZIP28 or bZIP17. NAC103 regulates genes important for cell survival during ER stress such as calreticulin, calnexin, protein disulfide isomerase, and ubiquitin conjugase 32,Citation36,Citation39 and engages in downstream signaling of SOG1, a master regulator of DNA damage caused by genotoxic stress.Citation40 On the other hand, NAC089 is known to induce the expression of genes that coordinate PCD and autophagy such as BAG6 and the MC5 metacaspase.Citation15,Citation41 NAC089 also activates genes required for PCD in response to the Tobacco mosaic virus or Cucumber mosaic virus infection.Citation42 While others reported NAC089 is a secondary activator downstream of bZIP60 and bZIP28, our recent investigations indicate that bZIP60 and bZIP17 also coordinate to regulate NAC089 in response to plant virus infection.Citation15,Citation36
Considering these layers of transcriptional responses that include regulation of PCD and autophagy, we conducted experiments to determine if these processes are factors in regulating TuMV or PlAMV infection. For example, AtBAG7 protein is a factor controlling PCD in response to ER stress. In atbag7 knockout plants, ER stress inducers such as heat and the N-glycosylation inhibitor tunicamycin accelerated cell death.Citation43 Inoculating atbag7 plants showed minimal change virus levels in the inoculated or systemic leaves as assayed by in fluorescence, suggesting that AtBAG7 control of PCD does not affect virus accumulation. BAG6 is another factor that is associated with a several cellular processes including proteasomal elimination of malformed proteins, autophagy, and basal immunity to Botrytis cinerea.Citation41,Citation44–46 Loss of function mutations in BAG6 leads to loss of B. cinerea resistance and inhibition of autophagy in plants. When considering ER stress-related autophagy, it is also worth noting that IRE1b and BiP engage in the regulation of heat-induced autophagy.Citation47 Furthermore, we looked for changes in ATG8 lipidation following TuMV-GFP or PlAMV-GFP infection, as a hallmark of autophagic activity in wild-type Col-0 and ire1a/ire1b, bzip60, bzip28, bzip17, nac089 and bag6 knock-out plants.Citation36 The accumulation of ATG8-PE was consistent across all lines suggesting that viral induction of the IRE1-dependent or independent pathways does not lead to changes in autophagic activity in the cell.
The ERQC plays a major role in virus infection
The UPR is a molecular signaling network that modulates many cellular activities including innate immunity and virus infection across eukaryotes. Viral protein production leads to significant ER stress and this can often activate the UPR. The UPR has been implicated in regulating titers of many vector-borne flaviviruses such as JEV, DENV, Tick-borne encephalitis virus, WNV and ZIKV.Citation26,Citation27,Citation31,Citation48 The mechanistic link between the proteins encoded by the flaviviruses, BiP, and UPR sensors is not yet clear. Regarding plant potyvirus and potexvirus infection, it remains an open question whether some genes that are upregulated by the signal transduction machinery are factors that are incoportated into the viral replication complexes, promote cell-to-cell movement, or aid virion assembly.Citation27,Citation28 Until now, research shows that an increased pool of BiP is important to stabilize protein folding and alleviate ER stress that could otherwise lead to PCD.Citation7,Citation33 In early experiments, the PVX TGB3 gene was introduced into the tobacco mosaic virus (TMV) vector for higher expression in N. benthamiana plants and this produced necrotic lesion and oxidative stress in the inoculated leaves. BiP overexpression was sufficient to alleviate the TGB3-induced necrosis and oxidative stress.Citation37,Citation49
More recent studies conducting using the Arabidopsis bzip60, bzip28, and bzip17 knockout plants suggest that one or more arm of the UPR is involved in restricting virus infection.Citation36,Citation37 Tauroursodeoxycholic acid (TUDCA) is a chemical chaperone that reduces ER stress in plants and mammals by reducing protein aggregates in the ER. TUDCA acts in mammalian cells to impinge on the signaling machinery affecting protein folding.Citation50,Citation51 Heat stress treatment to atbag7 knock-out Arabidopsis seedlings caused a significant number to die off, but a greater number of seedlings recovered after treatment with TUDCA.Citation43 This is one of several examples in the literature demonstrating that TUDCA offers protection to abiotic-induced ER stress in plants.Citation16,Citation47,Citation52 When WT Col-0 leaves were treated with TUDCA and inoculated with PlAMV-GFP, fluorescence was reduced indicating that viral infection was reduced.Citation36 These data suggest that the ERQC likely helps cells to cope with plant virus-induced ER stress and there maybe additional factors that also infection ().
Conclusion
The accumulation of studies using knockout mutants of the UPR pathway supports a model that the UPR suppresses or restricts potexvirus and potyvirus infection in Arabidopsis ().Citation36 Both potyvirus and potexviruses enhance the expression of BiP, the master regulator of the UPR that plays a key role in protection against severe ER stress. This potentially potentially leads to the PCD. The IRE1-bZIP60 pathway is preferentially activated by PlAMV, PVX, TuMV, and PVY. Additionally, the bZIP28 pathway of the UPR appears to play a role in restricting TuMV infection while the bZIP17 pathway restricts PlAMV infection. Although BiP is engaged by multiple branches of the UPR, potexviruses and potyviruses appear to preferentially influence distinct arms of the UPR. This may be due to different interactions between the TGB3 and 6K2 proteins with the ER-resident stress sensors or different engagements with BiP in activating each UPR sensor. Although the primary role of BiP is protein folding, we cannot rule out a possible role in virus replication, movement, or assembly. Furthermore, it is not clear if the coordinate actions of bZIP17 and bZIP60 for limiting PlAMV infection or the coordinate actions of bZIP28 and bZIP60 for limiting the TuMV infection is the sole result of upregulating BiP or if other unknown transcriptional targets are crucial for limiting virus infection. Further investigations are needed to identify key UPR factors that may be potential targets for developing gene-editing strategies or gene silencing strategies to control virus infection in plants. Given the level of conservation of the UPR machinery in plants and mammals, a plant genetic model presents a valuable opportunity to understand viral strategies modulating UPR across eukaryotes. This knowledge provides the basis on which novel specifically targeted therapeutic drugs can be developed in translational medicine.
Additional information
Funding
References
- Leborgne-Castel N, Jelitto-Van Dooren EP, Crofts AJ, Denecke J. Overexpression of BiP in tobacco alleviates endoplasmic reticulum stress. Plant Cell [Internet] 1999; 11:1–5. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=144191&tool=pmcentrez&rendertype=abstract.
- Pincus D, Chevalier MW, Aragón T, van Anken E, Vidal SE, El-Samad H, Walter P. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 2010;8:e1000415.
- Srivastava R, Deng Y, Shah S, Rao AG, Howell SH. Binding protein is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in arabidopsis. Plant Cell [Internet] 2013; 25:1416–1429. Available from] ;:. : http://www.plantcell.org/lookup/doi/10.1105/tpc.113.110684.
- Behnke J, Feige MJ, Hendershot LM. BiP and its nucleotide exchange factors Grp170 and Sil1: mechanisms of action and biological functions HHS public access. J Mol Biol [Internet] 2015. 427:1589–1608. [cited 2019 Jun 12]. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4356644/pdf/nihms664816.pdf.
- Vitale M, Bakunts A, Orsi A, Lari F, Tadé L, Danieli A, Rato C, Valetti C, Sitia R, Raimondi A, et al. Inadequate BiP availability defines endoplasmic reticulum stress. Elife. 2019;8:1–17.
- Cesaratto F, Sasset L, Myers MP, Re A, Petris G, Burrone OR. BiP/GRP78 mediates ERAD targeting of proteins produced by membrane-bound ribosomes stalled at the STOP-codon. J Mol Biol [Internet] 2019; 431:123–141. Available from: doi:https://doi.org/10.1016/j.jmb.2018.10.009.
- Verchot J. How does the stressed out ER find relief during virus infection? Curr Opin Virol. 2016;17.
- Kono N, Amin-Wetzel N, Ron D. Generic membrane-spanning features endow IRE1α with responsiveness to membrane aberrancy. Mol Biol Cell [Internet] 2017; 28:2318–2332. Available from: : https://www.molbiolcell.org/doi/10.1091/mbc.e17-03-0144.
- Verchot J. The ER quality control and ER associated degradation machineries are vital for viral pathogenesis. Front Plant Sci [Internet] 2014 5:66[cited 2019 Jun 17]. Available from: http://journal.frontiersin.org/article/10.3389/fpls.2014.00066/abstract.
- Huang S, Xing Y, Liu Y. Emerging roles for the ER stress sensor IRE1α in metabolic regulation and disease. J Biol Chem [Internet] 2019; 294:18726–18741. Available from: http://www.jbc.org/lookup/doi/10.1074/jbc.REV119.007036.
- Iwawaki T, Hosoda A, Okuda T, Kamigori Y, Nomura-Furuwatari C, Kimata Y, Tsuru A, Kohno K. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat Cell Biol. 2001;3:158–164.
- Bennett MK, Wallington-Beddoe CT, Pitson SM. Sphingolipids and the unfolded protein response. Biochim Biophys Acta [Internet] 2019;:1. doi:https://doi.org/10.1016/j.bbalip.2019.06.002.
- Oikawa D, Kimata Y, Kohno K, Iwawaki T. Activation of mammalian IRE1α upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp Cell Res. 2009;315:2496–2504.
- Kimata Y. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol Biol Cell. 2003;14:2559–2569.
- Yang ZT, Wang MJ, Le SL, Lu SJ, Bi DL, Le SL, Song ZT, Zhang SS, Zhou SF, Liu JX. The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet. 2014;10:e1004243.
- Zhang L, Chen H, Brandizzi F, Verchot J, Wang A. The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genet. 2015;11:e1005164.
- Takayanagi S, Fukuda R, Takeuchi Y, Tsukada S, Yoshida K Gene regulatory network of unfolded protein response genes in endoplasmic reticulum stress. Cell Stress Chaperones [Internet] 2013; 18:11–23. [cited 2020 Jul 3]. Available from: http://link.springer.com/10.1007/s12192-012-0351-5
- Kanemoto S, Kondo S, Ogata M, Murakami T, Urano F, Imaizumi K. XBP1 activates the transcription of its target genes via an ACGT core sequence under ER stress. Biochem Biophys Res Commun [Internet] 2005. 331:1146–1153. [[cited 2019 Jun 13]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15882996.
- Herath V, Gayral M, Adhikari N, Miller R, Verchot J Genome-wide identification and characterization of Solanum tuberosum BiP genes reveals the role of the promoter architecture in BiP gene diversity. [cited 2020 Jul 3]. Available from: https://doi.org/10.1101/2020.05.16.098244
- Angelos E, Ruberti C, Kim S-J, Brandizzi F. Maintaining the factory: the roles of the unfolded protein response in cellular homeostasis in plants. Plant J [Internet] 2017 90:671–682. [cited 2019 Jun 25]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27943485.
- Nawkar GM, Lee ES, Shelake RM, Park JH, Ryu SW, Kang CH, Lee SY. Activation of the transducers of unfolded protein response in plants. Front Plant Sci [Internet] 2018; 9:214. Available from: http://journal.frontiersin.org/article/10.3389/fpls.2018.00214/full.
- Li Y, Williams B, Dickman M. (BAG7)-mediated heat tolerance requires translocation, sumoylation and binding to WRKY29. New Phytol. 2017;2:695–705.
- Srivastava R, Deng Y, Howell SH. Stress sensing in plants by an ER stress sensor/transducer, bZIP28. Front Plant Sci. 2014;5:1–6.
- Henriquez-Valencia C, Moreno AA, Sandoval-Ibañez O, Mitina I, Blanco-Herrera F, Cifuentes-Esquivel N, Orellana A. bZIP17 and bZIP60 regulate the expression of BiP3 and other salt stress responsive genes in an UPR-independent manner in Arabidopsis thaliana. J Cell Biochem. 2015;116:1638–1645.
- Liu JX, Howell SH. bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell. 2010;22:782–796.
- Yu C-Y, Hsu Y-W, Liao C-L, Lin Y-L. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J Virol [Internet] 2006; 80:11868–11880. Available from: https://jvi.asm.org/content/80/23/11868.
- Peña J, Harris E. Dengue virus modulates the unfolded protein response in a time-dependent manner. J Biol Chem. 2011;286:14226–14236.
- Lewy TG, Grabowski JM, Bloom ME. BiP: master regulator of the unfolded protein response and crucial factor in flavivirus biology. Yale J Biol Med. 2017;90:291–300.
- Shi M, Lin X-D, Vasilakis N, Tian J-H, Li C-X, Chen L-J, Eastwood G, Diao X-N, Chen M-H, Chen X, et al. Divergent viruses discovered in arthropods and vertebrates revise the evolutionary history of the flaviviridae and related viruses. J Virol. 2016;90:659–669.
- Sharma M, Bhattacharyya S, Sharma KB, Chauhan S, Asthana S, Abdin MZ, Vrati S, Kalia M. Japanese encephalitis virus activates autophagy through XBP1 and ATF6 ER stress sensors in neuronal cells. J Gen Virol. 2017;98:1027–1039.
- Yu C, Achazi K, Niedrig M. Tick-borne encephalitis virus triggers inositol-requiring enzyme 1 (IRE1) and transcription factor 6 (ATF6) pathways of unfolded protein response. Virus Res. 2013;178: 471–477.
- Lu Y, Yin M, Wang X, Chen B, Yang X, Peng J, Zheng H, Zhao J, Lin L, Yu C, et al. The unfolded protein response and programmed cell death are induced by expression of Garlic virus X p11 in Nicotiana benthamiana. J Gen Virol. 2016;97:1462–1468.
- Williams B, Verchot J, Dickman MB. When supply does not meet demand-ER stress and plant programmed cell death. Front Plant Sci [Internet] 2014; 5:1–9. Available from: http://journal.frontiersin.org/article/10.3389/fpls.2014.00211/abstract.
- Verchot J. Cellular chaperones and folding enzymes are vital contributors to membrane bound replication and movement complexes during plant RNA virus infection. Front Plant Sci [Internet] 2012; 3:275.[cited 2019 Jun 16]. Available from: http://journal.frontiersin.org/article/10.3389/fpls.2012.00275/abstract.
- Gaguancela OA, Źuñiga LP, Arias AV, Halterman D, Flores FJ, Johansen IE, Wang A, Yamaji Y, Verchot J. The IRE1/bZIP60 pathway and bax inhibitor 1 suppress systemic accumulation of potyviruses and potexviruses in arabidopsis and nicotiana benthamiana plants. Mol Plant-Microbe Interact. 2016;29:750–766.
- Gayral M, Arias Gaguancela O, Vasquez E, Herath V, Flores FJ, Dickman MB, Verchot J. Multiple ER‐to‐nucleus stress signaling pathways are activated during Plantago asiatica mosaic virus and Turnip mosaic virus infection in Arabidopsis thaliana. Plant J [Internet] 2020:tpj.14798. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/tpj.14798
- Ye C, Dickman MB, Whitham SA, Payton M, Verchot J. The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol. 2011;156:741–755.
- Ye C, Verchot J. Role of unfolded protein response in plant virus infection. Plant Signal Behav [Internet] 2011; 6:1212–1215. Available from: http://www.tandfonline.com/doi/abs/10.4161/psb.6.8.16048.
- Sun L, Yang Z-T, Song Z-T, Wang M-J, Sun L, Lu S-J, Liu J-X. The plant-specific transcription factor gene NAC103 is induced by bZIP60 through a new cis -regulatory element to modulate the unfolded protein response in Arabidopsis. Plant J [Internet] 2013; 76:274–286. Available from: http://doi.wiley.com/10.1111/tpj.12287.
- Ryu TH, Go YS, Choi SH, Il KJ, Chung BY, Kim JH. SOG1-dependent NAC103 modulates the DNA damage response as a transcriptional regulator in Arabidopsis. Plant J. 2019;98:83–96.
- Li Y, Dickman M. Processing of AtBAG6 triggers autophagy and fungal resistance. Plant Signal Behav [Internet] 2016;11:e1175699. doi:https://doi.org/10.1080/15592324.2016.1175699.
- Li FF, Sun HJ, Jiao YB, Wang FL, Yang JG, Shen LL. Viral infection-induced endoplasmic reticulum stress and a membrane-associated transcription factor NbNAC089 are involved in resistance to virus in Nicotiana benthamiana. Plant Path. 2018;67:233–243.
- Williams B, Kabbage M, Britt R, Dickman MB. AtBAG7, an arabidopsis Bcl-2-associated athanogene, resides in the endoplasmic reticulum and is involved in the unfolded protein response. Proc Natl Acad Sci USA [Internet] 2010; 107:6088–6093. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20231441.
- Kawahara H, Minami R, Yokota N. BAG6/BAT3: emerging roles in quality control for nascent polypeptides. J Biochem. 2013;153:147–160.
- Yamamoto K, Hayashishita M, Minami S, Suzuki K, Hagiwara T, Noguchi A, Kawahara H. Elimination of a signal sequence-uncleaved form of defective HLA protein through BAG6. Sci Rep. 2017;7:14545.
- Li Y, Kabbage M, Liu W, Dickman MB. Aspartyl protease-mediated cleavage of BAG6 is necessary for autophagy and fungal resistance in plants. Plant Cell [Internet] 2016; 28:233–247. Available from: http://www.plantcell.org/lookup/doi/10.1105/tpc.15.00626.
- Yang X, Srivastava R, Howell SH, Bassham DC. Activation of autophagy by unfolded proteins during endoplasmic reticulum stress. Plant J. 2016;85:83–95.
- Tan Z, Zhang W, Sun J, Fu Z, Ke X, Zheng C, Zhang Y, Li P, Liu Y, Hu Q, et al. ZIKV infection activates the IRE1-XBP1 and ATF6 pathways of unfolded protein response in neural cells. J Neuroinflammation. 2018;15:1–16.
- YE C-M, Chen S, Payton M, Dickman MB, Verchot J. TGBp3 triggers the unfolded protein response and SKP1-dependent programmed cell death. Mol Plant Pathol [Internet] 2013; 14:241–255. doi: https://doi.org/10.1111/mpp.12000.
- Uppala JK, Gani AR, Ramaiah KVA. Chemical chaperone, TUDCA unlike PBA, mitigates protein aggregation efficiently and resists ER and non-ER stress induced HepG2 cell death. Sci Rep [Internet] 2017 ; 7:3831. [cited 2019 Jun 26]. Available from]: http://www.nature.com/articles/s41598-017-03940-1.
- Fernández-Bautista N, Fernández-Calvino L, Muñoz A, Castellano MM. HOP3, a member of the HOP family in Arabidopsis, interacts with BiP and plays a major role in the ER stress response. Plant Cell Environ. 2017;40: 1341–1355.
- Ishikawa T, Watanabe N, Nagano M, Kawai-Yamada M, Lam E. Bax inhibitor-1: a highly conserved endoplasmic reticulum-resident cell death suppressor. Cell Death Differ. 2011;18:1271–1278.
