ABSTRACT
Calcium-dependent protein kinases-related kinases (CDPK-related kinases; CRKs) are Ser/Thr kinases that bind with Ca2+/Calmodulin and play crucial roles in signal transduction pathways during plant growth, development, and responses to multiple stresses. In this study, we have studied detailed organ and tissue-specific expression patterns of rice CRK genes. Our organ-specific RT-PCR analyzes show the differential expression pattern of these genes in various organs of rice. Moreover, our RNA-RNA in situ hybridization study in rice stem base containing developing crown root primordia demonstrates that the expression of CRK genes is spatially restricted to the developing crown root primordia, suggesting their putative role in protein phosphorylation-dependent cellular signaling during rice crown root development. Furthermore, organ-specific differentially expression pattern of CRK genes during floral organogenesis further support for the organ-specific cell signaling during organogenesis. Thus, our study provides a developmentally regulated expression pattern of rice CRK genes, though they are broadly expressed and a basic foundation for functional characterizations of CRK gene members to unravel their specific functions during plant growth and development.
Introduction
Plants have developed a sophisticated signal transduction pathway to adapt for climate change. Calcium (Ca2+) plays a vital role in signal transduction as a secondary messenger during various biological processes including growth and development.Citation1 Transient fluctuations in Ca2+ concentrations in the cytoplasm can be sensed by members of four kinases superfamilies whose activities are regulated by calcium and/or calmodulin (CaM). These are Ca2+-dependent protein kinases (CDPKs), Ca2+/calmodulin-dependent protein kinases (CaMKs), calcium and calmodulin-dependent protein kinases (CCaMKs) and CDPK-related protein kinases (CRKs).Citation2–5 CDPK-related kinases (CRKs) basically belong to a Ser/Thr protein kinase group while some of the AtCRKs have Tyr kinase activity but they are closely related to CDPKs.Citation6 They possess a CDPK-related catalytic domain and a C-terminal CaM domain-containing degenerated EF-hand Ca2+ binding motifs.Citation7,Citation8 Members of CDPKs/CRKs superfamilies contain N-terminal myristoylation sites that target them to get localized in the plasma membrane and endomembrane system.Citation9 Unlike CDPKs, plant CRKs are not well characterized. A few studies in dicot plants have shown their role in plant development and stress toleranceCitation10-14 but they are not studied in monocot plants, particularly in crop species. In Arabidopsis, AtCRK1 positively regulates heat and salt stresses in the plant whereas AtCRK3 controls the remobilization of nitrogen during leaf senescence.Citation11,Citation12 Importantly, a plasma membrane-localized AtCRK5 is required for proper root growth, gravity responses, embryogenesis, and hypocotyls hook formation during skotomorphogenesis.Citation13,Citation15,Citation16 Its inactivation results in the inhibition of the root growth, enhanced lateral root formation, and defective gravitism in Arabidopsis due to defect in polar auxin distribution, strongly suggesting functions of CRKs in the regulation of hormonal signaling during plant growth and development.Citation9,Citation13 Similarly, tomato LeCRK1 has an important role in the fruit ripening process.Citation10 In rice, interestingly OsCBK (OsCRK1) binds to calmodulin in a calcium-dependent manner but its enzymatic activity is independent of calcium and calmodulin, indicating that its interaction with Ca2+/calmodulin may more relevant for the regulation and may not be crucial for its direct kinase function.Citation17
Here, we have studied the expression pattern of five predicted rice CRK genes in various organs. We show the differential expression levels of these genes in different organs. We reveal the spatial transcript distribution pattern of these genes in the stem base (crown tissue) which contains developing crown root primordia at different developmental stages and during floral organogenesis using RNA-RNA in-situ hybridization. Our results provide a base for detailed functional characterization of rice candidate CRK genes for revealing their tissue-specific roles during growth and development.
Materials and methods
Plant material and growth condition
Rice (Oryza sativa subsp. indica cv. IR64) seeds were germinated on ½ MS media and grown in a controlled condition at 27°C with a 16/8 h light/dark daily cycle as described earlier.Citation18 For organ-specific expression analysis, stem base (about 2 mm), crown root, primary root, leaf blade, and whole seedling were collected from 6d old rice plants, while 0.5–1.0 cm panicles were collected from the growing rice plants.
Expression analysis by RT-PCR
For reverse transcription-polymerase chain reaction (RT-PCR), total RNAs from seedling, crown tissue (stem base), crown root, primary root, leaf blade, and panicles were extracted using TRI reagent (Sigma, Missouri, USA). The RNAs were quantified using nanodrop (Thermo Fisher Scientific, Massachusetts, USA) and the quality of RNA samples was ensured by running total RNAs on 1% agarose gel. For reverse transcription, 1 μg total RNAs were used for first-strand cDNA synthesis using the iScript cDNA synthesis kit (BioRad, California, USA). Gene-specific primers for each gene were designed by PrimerQuest Tool (https://eu.idtdna.com/PrimerQuest/Home/Index) and rice Ubq5 gene (GenBank accession number: AK061988) was used as an internal control (Supplementary Table 1). The relative expression level was calculated by quantifying band intensity and normalizing with an internal control using Image Lab Software (BioRad, California, USA). Data were depicted as mean value ±SD of three independent biological replicates. The data were analyzed by one-way analysis of variance (ANOVA) at (P ≤ 0.05) using SPSS v.16.0 (SPSS, Chicago, USA). Statistical analysis and column diagrams have been prepared by using Prism 8, Graphpad software (https://www.graphpad.com). Data obtained from semi-quantitative-PCR in three independent biological replicates were used to produce heatmap through Bioconductor R (http://www.bioconductor.org).
RNA-RNA in situ hybridization
For RNA-RNA in situ hybridization analysis, as described previously by Neogy et al., (2019),Citation18 a rice stem base and young panicles undergoing floret organogenesis were fixed in FAA (10% formaldehyde, 5% glacial acetic acid and 50% ethanol) fixative, dehydrated through ethanol-xylene series and embedded in paraffin (Sigma, Missouri, USA). An 8 µm cross-sections of stem base and longitudinal sections of panicles were made using a microtome (Thermo Fisher Scientific, Massachusetts, USA) and collected on poly-L-lysine coated slides. For generating RNA probes, 110 to 130-bp gene-specific regions of CRK genes were PCR amplified using cDNA (Supplementary Table 1) and were cloned into pBluescript SK+ vector as a blunt in EcoRV restriction site. The orientation of the fragments for T7/T3 promoters has verified the sequencing. OsCRK1, OsCRK3, and OsCRK4 genes were cloned in anti-sense direction while OsCRK2 and OsCRK5 genes were cloned in the sense direction. The antisense probe for OsCRK1, OsCRK3, and OsCRK4 was transcribed with T7 RNA Polymerase (Sigma-Aldrich, USA) using EcoRI linearized pBluescript SK+ clones. However, the sense probe for OsCRK2 and OsCRK5 were transcribed with T3 RNA Polymerase (NEB, USA) using HindIII linearized pBluescript SK+ clones. Slides were treated and hybridized as described previously.Citation19,Citation20 A color signal was developed performing enzymatic reaction for alkaline phosphatase (AP) which is conjugated with anti-DIG antibodies (Sigma-Aldrich, USA) using NBT/BCIP substrate (Sigma-Aldrich, USA) for the reaction (Sigma-Aldrich, USA). Entellan (Merck- Millipore, Germany) mounted sections were imaged under a bright-field microscope with a device camera (Carl Zeiss AG, Germany).
Results and discussion
Protein phosphorylation and dephosphorylation provide a key post-translational regulatory switch for regulating conformational changes, sub-cellular localization, and molecular interactions of proteins during several fundamental biological processes including growth and development. For example, protein phosphorylation is important in asymmetric localization of PIN proteins required for polar auxin transport during plant growth and development.Citation13,Citation21,Citation22 Similarly, phosphorylation is a crucial event for two-component cytokinin signaling in plants.Citation23 In Arabidopsis, AtCRK5 has been shown to play a crucial role during various stages of plant growth and development.Citation9,Citation13,Citation15,Citation16 AtCRK5 is specifically required for proper PIN2 localization in the transition zone of the primary root.Citation13 It also controls auxin-gibberellic acid cross-talk during embryogenesis and auxin-ethylene-gibberellic acid crosstalk during hypocotyl hook development during skotomorphogenesis.Citation15,Citation16
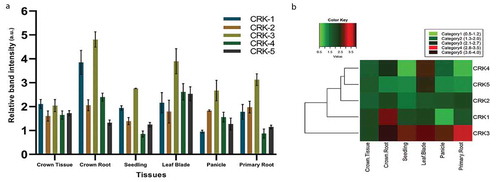
Toward investigating functions of CRK genes during growth and development in rice, we investigated expression patterns of five putative rice CRK genes in six tissues. We performed RT-PCR analysis for these genes using RNAs from rice crown tissues at the stem base, emerged crown root, seedling, leaf blade, early panicle, and primary root. Our analysis showed that CRK genes were broadly expressed in all these rice tissues ()) but transcript abundance is differential across these tissues. The expression level of OsCRK1 is highest in the emerged crown roots and lowest in the panicles (,b)). Similarly, OsCRK3 is also most abundant in the crown roots but minimum in the crown tissues (,b)). On the other hand, the expression level of OsCRK4 and OsCRK5 was maximum in the leaf blades (,b)). Interestingly, the expression levels of OsCRK1, OsCRK3, and OsCRK4 in primary roots are lower than the crown roots, suggesting root-type differential expression of these genes. However, we do not observe much difference in the expression level of OsCRK2 across these tissues. A complete linkage algorithm and a euclidean distance matrix were used to construct a heat map by clustering of expressed genes for the different tissues ()). The clustering tree is showing the division of genes into two broad groups; group 1 comprising of OsCRK1, 2, 4, 5, and the second group with only OsCRK3 ()). Based on this expression pattern, OsCRK1, 3, and 4 may have differential functions between primary and crown roots ()).
Figure 1. Organ-specific expression pattern of rice CRK genes. (a) Expression levels based on relative band intensity of CRK genes in different tissue of rice. (b) Heatmap analysis of CRK genes in different tissue. This figure combines a “heatmap” with a dendrogram. The rows represent genes while the type of tissues is shown in the columns. The expression levels are mapped on the color scale with low, intermediate, and high expression represented by green, red, and black color respectively
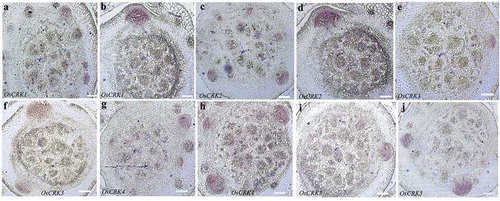
Since the expression of some of the CRK genes was lower in the stem base containing developing crown root primordia as compared with emerged crown roots, we analyzed their spatial expression pattern in the internal tissues of the stem base. RNA in situ hybridization was performed on the cross-sections of the stem base using antisense RNA probes for each gene. Important to note that the expression of all these CRK genes was mostly restricted to the developing crown root primordia in the stem base and no expression above the background was observed in other tissue ()). The CRK genes are activated in the initiating crown root primordia and continue to express during outgrowth of the primordia ()). These observations together with the RT-PCR data suggest that rice CRK genes are expressed since the early stages of the crown root primordia and continue to express in the growing crown roots, post-emergence of the crown root primordia.
Figure 2. Spatial expression pattern of CRK genes during rice crown root primordia development. (aj) Tissue-specific expression pattern of OsCRK1 (a,b), OsCRK2 (c,d), OsCRK3 (e,f), OsCRK4 (g,h), and OsCRK5 (i,j) during CRP development, hybridized with anti-sense DIG-RNA probe. Bars: 20 µm
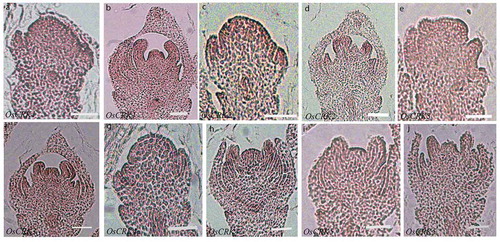
Similarly, we also studied the spatial expression pattern of rice CRK genes during rice floret development. Young panicle (0.5–1 cm) containing florets undergoing different stages of floral organogenesis were sectioned longitudinally and hybridized with DIG-UTP labeled anti-sense RNA probes against all five CRK genes. We observed that expression of OsCRK1 and OsCRK2 was relatively higher in the floret meristem as compared to OsCRK3-5 (,,,,)). They are expressed throughout the floret meristem including developing lemma and palea but during a later stage of floret organogenesis, their expression is restricted to the inner floret organs and is significantly decreased in the lemma and palea ()). OsCRK1, OsCRK3, and OsCRK4 are expressed in developing lodicules, stamens, and carpel (,,)) whereas OsCRK2 has relatively stronger expression in lodicules and stamens as compared to carpel tissues ()). However, the expression of OsCRK5 is not significantly higher than the background level ()). All these data together suggest that rice CRK genes display tissue and organ-specific differential expression pattern.
Figure 3. Spatial expression pattern of CRK genes during rice floret development
Conclusion
In the present study, our data showed that though all five rice CRK genes are expressed broadly but they display differential expression levels in different organs, suggesting their organ-specific regulation. This also suggests that they might have specific functions in different organs/tissues. Moreover, spatially regulated expression of these genes in the stem base and floret organs further supports toward their tissue-specific differential functions during organ growth and development. The functions of these CRKs remain elusive and this work can provide a foundation for the further functional characterization of these genes in rice as well as in other plant species.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental Material
Download PDF (5.9 KB)Acknowledgments
This work was supported by the Science and education research board (SERB), Government of India under National Postdoctoral Fellowship (File No: PDF/2016/002194) to A. Y. The Department of Biotechnology, Indian Institute of Technology, Roorkee, India is acknowledged for providing infrastructure and administrative supports. Indian Institute of Technology, Roorkee, India, and University Grant Commission (UGC), India are acknowledged for providing fellowships to T.G. and H.S., respectively.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
References
- Hepler PK. Calcium: a central regulator of plant growth and development. Plant Cell. 2005;17:1–4. doi:https://doi.org/10.1105/tpc.105.032508.
- Zuo R, Hu R, Chai G, Xu M, Qi G, Kong Y, Zhou G. Genome-wide identification, classification, and expression analysis of CDPK and its closely related gene families in poplar (Populus trichocarpa). Mol Biol Rep. 2013;40:2645–2662. doi:https://doi.org/10.1007/s11033-012-2351-z.
- Cai H, Cheng J, Yan Y, Xiao Z, Li J, Mou S, Qiu A, Lai Y, Guan D, He S. Genome-wide identification and expression analysis of calcium-dependent protein kinase and its closely related kinase genes in Capsicum annuum. Front Plant Sci. 2015;6:737. doi:https://doi.org/10.3389/fpls.2015.00737.
- Wang JP, Xu YP, Munyampundu JP, Liu TY, Cai XZ. Calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in tomato: genome-wide identification and functional analyses in disease resistance. Mol Genet Genomics. 2016;291:661–676. doi:https://doi.org/10.1007/s00438-015-1137-0.
- Zhang H, Wei C, Yang X, Chen H, Yang Y, Mo Y, Li H, Zhang Y, Ma J, Yang J, et al. Genome-wide identification and expression analysis of calcium-dependent protein kinase and its related kinase gene families in melon (Cucumis melo L.). PloS One. 2017;12:e0176352. doi:https://doi.org/10.1371/journal.pone.0176352.
- Nemoto K, Takemori N, Seki M, Shinozaki K, Sawasaki T. Members of the plant CRK superfamily are capable of trans- and autophosphorylation of tyrosine residues. J Biol Chem. 2015;290:16665–16677. doi:https://doi.org/10.1074/jbc.M114.617274.
- Furumoto T, Ogawa N, Hata S, Izui K. Plant calcium-dependent protein kinase-related kinases (CRKs) do not require calcium for their activities. FEBS Lett. 1996;396:147–151. doi:https://doi.org/10.1016/0014-5793(96)01090-3.
- Harmon AC, Gribskov M, Harper JF. CDPKs-a kinase for every Ca2+ signal? Trends Plant Sci. 2000;5:154–159. doi:https://doi.org/10.1016/s1360-1385(00)01577-6.
- Baba AI, Rigo G, Ayaydin F, Rehman AU, Andrasi N, Zsigmond L, Valkai I, Urbancsok J, Vass I, Pasternak T, et al. Functional analysis of the Arabidopsis thaliana CDPK-related kinase family: atCRK1 regulates responses to continuous light. Int J Mol Sci. 2018;19:1282. doi:https://doi.org/10.3390/ijms19051282.
- Leclercq J, Ranty B, Sanchez-Ballesta MT, Li ZG, Jones B, Jauneau A, Pech JC, Latche A, Ranjeva R, Bouzayen M. Molecular and biochemical characterization of LeCRK1, a ripening associated tomato CDPK-related kinase. J Exp Bot. 2005;56:25–35. doi:https://doi.org/10.1093/jxb/eri003.
- Li RJ, Hua W, Lu YT. Arabidopsis cytosolic glutamine synthetase AtGLN1;1 is a potential substrate of AtCRK3 involved in leaf senescence. Biochem Bioph Res Co. 2006;342:119–126. doi:https://doi.org/10.1016/j.bbrc.2006.01.100.
- Liu HT, Gao F, Li GL, Han JL, Liu DL, Sun DY, Zhou RG. The calmodulin-binding protein kinase 3 is part of heat-shock signal transduction in Arabidopsis thaliana. Plant J. 2008;55:760–773. doi:https://doi.org/10.1111/j.1365-313X.2008.03544.x.
- Rigo G, Ayaydin F, Tietz O, Zsigmond L, Kovacs H, Pay A, Salchert K, Darula Z, Medzihradszky KF, Szabados L, et al. Inactivation of plasma membrane-localized CDPK-RELATED KINASE 5 decelerates PIN2 exocytosis and root gravitropic response in Arabidopsis. Plant Cell. 2013;25:1592–1608. doi:https://doi.org/10.1105/tpc.113.110452.
- Tao XC, Lu YT. Loss of AtCRK1 gene function in Arabidopsis thaliana decreases tolerance to salt. J Plant Biol. 2013;56:306–314. doi:https://doi.org/10.1007/s12374-012-0352-z.
- Baba AI, Andrasi N, Valkai I, Gorcsa T, Koczka L, Darula Z, Medzihradszky KF, Szabados L, Feher A, Rigo G, et al. AtCRK5 protein kinase exhibits a regulatory role in hypocotyl hook development during skotomorphogenesis. Int J Mol Sci. 2019a;20:3432. doi:https://doi.org/10.3390/ijms20143432.
- Baba AI, Valkai I, Labhane NM, Koczka L, Andrasi N, Klement E, Darula Z, Medzihradszky KF, Szabados L, Feher A, et al. CRK5 protein kinase contributes to the progression of embryogenesis of Arabidopsis thaliana. Int J Mol Sci. 2019b;20:6120. doi:https://doi.org/10.3390/ijms20246120.
- Zhang L, Bi-Feng LIU, Liang S, Jones RL, Ying-Tang LU. Molecular and biochemical characterization of a calcium/calmodulin-binding protein kinase from rice. Biochem J. 2002;368:145–157. doi:https://doi.org/10.1042/bj20020780.
- Neogy A, Garg T, Kumar A, Dwivedi AK, Singh H, Singh U, Singh Z, Prasad K, Jain M, Yadav SR. Genome-wide transcript profiling reveals an auxin-responsive transcription factor, OsAP2/ERF-40, promoting rice adventitious root development. Plant Cell Physiol. 2019;60:2343–2355. doi:https://doi.org/10.1093/pcp/pcz132.
- Prasad K, Parameswaran S, Vijayraghavan U. OsMADS1, a rice MADS box factor, controls differentiation of specific cell types in the lemma and palea and is an early acting regulator of inner floral organs. Plant J. 2005;43:915–928. doi:https://doi.org/10.1111/j.1365-313X.2005.02504.x.
- Yadav SR, Prasad K, Vijayraghavan U. Divergent regulatory OsMADS2 functions control size, shape and differentiation of the highly derived rice floret second-whorl organ. Genetics. 2007;176:283–294. doi:https://doi.org/10.1534/genetics.107.071746.
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130:1044–1056. doi:https://doi.org/10.1016/j.cell.2007.07.033.
- Barbosa IC, Hammes UZ, Schwechheimer C. Activation and polarity control of PIN-FORMED auxin transporters by phosphorylation. Trends Plant Sci. 2018;23:523–538. doi:https://doi.org/10.1016/j.tplants.2018.03.009.
- Du L, Jiao F, Chu J, Jin G, Chen M, Wu P. The two-component signal system in rice (Oryza sativa L.): a genome-wide study of cytokinin signal perception and transduction. Genomics. 2007;89:697–707. doi:https://doi.org/10.1016/j.ygeno.2007.02.001.
