ABSTRACT
Mitogen-activated protein kinase kinase kinases (MAPKKKs or MEKKs) are crucial components of the MAPK signaling cascades, which play central roles in the signaling transduction pathways for plant growth, development, and response to abiotic stresses such as drought. The MAPKKK gene families in pepper have not been functionally characterized yet. Here, from the pepper genome, we predicted 27 putative MAPKKK genes belonging to the MEKK subfamily (named CaMEKK1-27), based on in silico analysis. Phylogenetic analysis revealed that 14 CaMEKK genes were clustered into A5 of the five groups (A1-A5), of which 9 genes are primarily on chromosomes 2 and 7, and are located close to each other. These nine genes showed transcriptional regulation by treatment with abscisic acid (ABA) and drought stress in quantitative reverse-transcription PCR analysis. Among the ABA- and/or drought-induced CaMEKK genes, in a previous study, we isolated CaAIMK1 (Capsicum annuum ABA Induced MAP Kinase 1), which plays a positive role in drought resistance via an ABA-dependent pathway. Our expression analysis and functional characterization of the MEKK subfamily genes will provide a better understanding of the functional roles of pepper MAPK cascades in ABA-mediated drought responses.
KEYWORDS:
As highly conserved signaling modules in eukaryotes, mitogen-activated protein kinase (MAPK) cascades play crucial roles in signal transduction in various plant biological processes, including growth, development, senescence, responses to hormones, and regulation of biotic and abiotic stresses.1–Citation5 MAPK cascades typically consist of at least three functionally linked serine/threonine kinases: a MAPKKK (also known as MAP3Ks or MEKKs), a MAPKK (also known as MKK/MEK), and a MAPK (also known as MPKs).Citation3,Citation6 When receptors receive an extracellular stimulus, a sequential activation of MAPK cascades through phosphorylation events activates downstream signals such as the induction of transcription factors and enzymes in response to various environmental stimuli.Citation7,Citation8 In the Arabidopsis genome, 20 MAPKs, 10 MAPKKs, and 80 MAPKKKs have been identified.Citation3,Citation6 Similar gene sets have been evolutionarily conserved among higher plants, although there are variations in gene numbers within a plant species.Citation2,Citation9–11 MPK3, MPK4, and MPK6 are components of the Arabidopsis MAPK cascades, which are well known for being involved in plant responses to various stimuli, including pathogen-associated molecular patterns, H2O2, cold, salt, osmotic shock, and hormones such as ABA, ethylene, and jasmonic acid.Citation6,Citation12,Citation13 MAPKKKs, which are the largest group compared to MAPKKs and MAPKs, function as adapters connecting the upstream signal step to the MAPK cascade. However, a few MAPKKKs have been functionally characterized. Arabidopsis MAPKKKs can be divided into three subfamilies: MEKK (21 members), Raf (48 members), and ZIK (11 members).Citation9,Citation14 Several studies have shown the activation of MAPK cascades in an ABA-dependent manner in various plant species, including Arabidopsis, tobacco, pea, maize, and rice.Citation3,Citation9 In particular, many genes of Arabidopsis MEKK subfamily, including MAPKKK1 (ANP1), MAPKKK10 (MEKK3), MAPKKK14, MAPKKK15, MAPKKK16, MAPKKK17, MAPKKK18, and MAPKKK19, were transcriptionally regulated by treatment with ABA.Citation8,Citation15,Citation16 In response to ABA, MAPKKK17/18 phosphorylates MKK3 and consequently activates the downstream MAPKs MPK1, MPK2, MPK7, and MPK14 proteins. In this process, it has been proposed that the MAPKKK17/18-MKK3-MPK1/2/7/14 cascade is regulated at transcriptional and translational levels by the ABA core signaling module Pyrabactin Resistance (PYR)/PYR-Like (PYL)/Regulatory Component of ABA Receptor (RCAR)-Type 2 C Protein Phosphatase (PP2C)- SNF1-Related Protein Kinase 2 (SnRK2).Citation5,Citation7,Citation8 Similar studies have been conducted in several other plant species, such as rice, maize, and tomato.Citation10,Citation17–19 In pepper, Li et al. (2012)Citation20 have identified 19 MPK genes and 5 MKK genes, and Iftikhar et al. (2017)Citation10 predicted 14 MAPKs, 5 MKKs, and 60 MAPKKKs (17 in MEKK subfamily, 37 in the Raf subfamily, and 6 in the ZIK subfamily); however, information on the functional involvement of those genes, in particular MEKK subfamily genes, in response to ABA signaling and drought responses is still lacking.
Many genes encoding the component of a MAPK signaling cascade have been identified from several plant species, including Arabidopsis,Citation3,Citation6 rice,Citation19 tomato,Citation10,Citation18 and maize,Citation17 and a similar number of gene sets in higher plants indicate evolutionary conservation of MAPK cascade genes.Citation9 Based on these, we initially collected full-length amino acid sequences of 21 and 17 MEKK subfamily genes from the model plant Arabidopsis, and also from tomato belonging to the same Solanaceae family as pepper. We used those sequences as a query in a BLASTP search to retrieve pepper gene sequences encoding MEKK subfamily genes involved in ABA and drought-stress signaling. Among the pepper orthologous genes, 27 genes were selected as putative MEKK genes and named CaMEKK1-27 (). These genes encode the protein kinase with a Ser/Thr kinase domain, and also have a conserved kinase domain of G(T/S)Px(W/Y/F)MAPEV found in the MEKK subfamily proteins,Citation14 although there are some mutated residues. Subcellular localization analysis predicted that the CaMEKK proteins are mainly localized to the nucleus. Using the full-length amino acid sequences of the MEKK genes from pepper, Arabidopsis, and tomato, we conducted multiple sequence alignment and phylogenetic analysis using the neighbor-joining method with MEGA X software.Citation21 In the phylogenetic tree, the MEKK genes were clustered into five clades and were grouped (A1-A5) according to the results of previous studiesCitation10,Citation22 ( and ). Of the five groups, none of the pepper genes belonged to group A1, while group A5 is the largest with 14 pepper genes.
Table 1. List of CaMEKK genes and their related information in the pepper plant
Figure 1. Phylogenetic tree analysis of putative MEKK subfamily genes in pepper. The amino acid sequences were deduced from the full-length coding sequences of MEKK genes in pepper (green diamonds), Arabidopsis (red triangles), and tomato (orange circles), and were used for comparison. Multiple alignment analysis was performed using ClustalW and the phylogenetic tree was drawn using the neighbor-joining method with MEGA X software.Citation21 Bootstrap values were calculated from 1,000 bootstrap replications and are indicated at each branch point. Scale bar indicates the evolutionary distance computed using the Poisson correction method
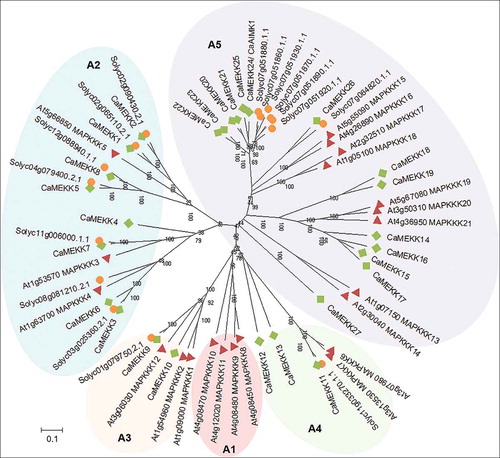
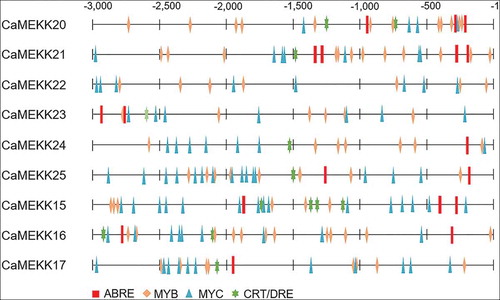
All the pepper MEKK genes, except CaMEKK27, in group A5 share sequence homology with Arabidopsis MAPKKK15-21. The transcriptional regulation of AtMAPKKK15–20 is enhanced by ABA.Citation15,Citation16,Citation23 In particular, it has been reported that AtMAPKKK18 plays a positive role in drought resistance by enhancing stomatal closure,Citation7,Citation24 and AtMAPKKK20 is functionally involved in ABA-induced stomatal closure.Citation23 AtMAPKKK18 is likely to be modulated by interaction with core components of ABA signaling, including protein phosphatase type 2 C (PP2C) ABI1Citation7 and the kinase SnRK2.6.Citation25 In the absence of ABA, AtMAPKKK18 is dephosphorylated by ABI1 and loses its kinase activity.Citation7 The inactivated AtMAPKKK18 protein is degraded by the ubiquitin-proteasome pathway, which is blocked in the presence of ABA, leading to the activation of downstream components.Citation7,Citation26 Based on the sequence homology, AtMAPKKK18 is closest in the order of CaMEKK25, CaMEKK24, CaMEKK20, CaMEKK22, CaMEKK23, CaMEKK21, CaMEKK26, and CaMEKK27 among the pepper MEKK genes, while AtMAPKKK20 is close to CaMEKK14, CaMEKK15, CaMEKK16, CaMEKK17, and CaMEKK18 in order. Interestingly, 9 of these genes are located close to each other on the chromosome: CaMEKK15, CaMEKK16, and CaMEKK17 on chromosome 2 (within 34.7 kilobase pair(kb)), and CaMEKK20, CaMEKK21, CaMEKK22, CaMEKK23, CaMEKK24, and CaMEKK25 on chromosome 7 (within 388.7 kb). Based on these, we finally selected the nine clustered genes in this study and analyzed whether the transcriptional induction of the CaMEKK genes can be controlled by drought stress and ABA. To do this, we first performed quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) analysis with RNAs from the third and fourth leaves from six-leaf stage pepper plants as described previouslyCitation11 (). cDNA was synthesized from 1 μg total RNA using a Transcript First Strand cDNA Synthesis kit (Roche, Indianapolis, IN, USA), according to the manufacturer’s instructions. For drought stress, pepper plants were carefully removed from the soil to prevent injury and then dried on 3 MM paper (Whatman) for 2 and 6 hours. As an Arabidopsis RD29B homolog, CaOSR1 gene induced by drought stress and ABACitation28 was used as a positive control for treatment. In response to drought stress, transcripts of the CaOSR1 gene were gradually accumulated, and of the CaMEKK genes, CaMEKK20 and CaMEKK25 showed relatively high induction levels (). The expression levels of CaMEKK genes, except for CaMEKK16 and CaMEKK17, significantly increased 2 h after treatment, compared to those of the control (0 h), and CaMEKK20, CaMEKK21, CaMEKK22, and CaMEKK23 genes showed continuously high expression at 6 h (). Since drought stress promotes ABA biosynthesis in plants, especially leaves,Citation29,Citation30 we also examined the expression patterns of CaMEKKs in response to ABA. ABA solution (100 μM) was foliar sprayed on the same stage plant as above and the leaves were harvested for RNA isolation 6 and 12 h after treatment. qRT-PCR analysis revealed that ABA treatment also regulated the expression of CaMEKK genes (). Following ABA treatment, CaMEKK24, CaMEKK25, and CaMEKK15 genes showed distinct induction, and the expression of CaMEKK20, CaMEKK22, and CaMEKK16 genes were highly induced 6 h or 12 h after treatment. Compared to drought stress, ABA-induced expression of CaMEKK genes was quite different; for example, highly induced CaMEKK20 by drought did not show induction after ABA treatment, while CaMEKK15 gene was induced by ABA, not drought stress. This discrepancy may be explained by the possibility of ABA-independent induction of the CaMEKK genes in response to drought stress. To further understand drought stress- and ABA-induced transcription regulation of the nine CaMEKK genes, we analyzed cis-regulatory elements, closely correlating with gene expression responses to stimuli, in their promoter sequences. Using the PLACE database (http://www.dna.affrc.go.jp/PLACE/)Citation31, cis-elements were predicted in the 3 kb upstream region of the CaMEKK genes. We found many of ABA- and drought stress-responsive cis-elements, such as ABA-responsive element (ABRE), MYB, MYC, and C-repeat/dehydration-responsive element (CRT/DRE) (). In particular, there was at least one ABRE in all the nine CaMEKK genes, and CaMEKK21 had the most ABREs with four. This prediction was not consistent with the expression patterns of the CaMEKK genes in response to ABA (). The data raised the following possibilities: (1) CaMEKK genes may be induced outside the time points tested in this study. (2) ABA concentration used in this study may not be sufficient for CaMEKK gene expression. (3) there may be other factors responsible for the ABA-induced expression of the CaMEKK genes.
Figure 2. Expression pattern of drought-induced MEKK genes in response to drought stress and ABA. Expression levels of CaMEKK genes were measured by qRT-PCR analysis using cDNA derived from total RNA that was extracted from the third and fourth leaves of six-leaf stage pepper plants following drought stress (a) and spraying with 100 μM ABA (b). Expression level of each CaMEKK gene was normalized to that of CaActin1, an internal control gene, calculated using the delta-delta Ct method,Citation27 and the value at 0 h was set to 1.0. qRT-PCR analysis was performed in duplicate, with three biological repeats, and data represent the mean ± standard error. Asterisks indicate significant differences compared to the value at 0 h of each gene (Student’s t-test; P < .05)
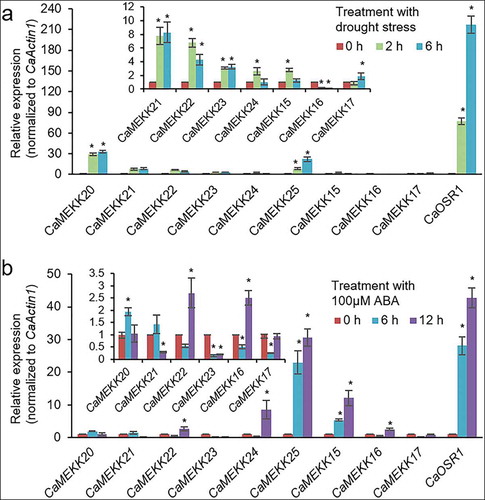
Figure 3. In silico analysis of cis-regulatory elements, associated with ABA signaling and drought stress, in the −3-kb upstream region of the CaMEKK genes. cis-regulatory elements were predicted using a web tool New PLACE (https://www.dna.affrc.go.jp/PLACE/?action=newplace) and were presented as different shapes with different colors. ABRE, ABA-responsive element; CRT/DRE, C-repeat/dehydration-responsive element
Based on our results, we expected that the nine pepper MEKK genes could be involved in drought stress response and/or ABA signaling. To test this hypothesis, we are investigating the functional role of these CaMEKK genes under abiotic stress conditions. As an example, we recently reported that CaAIMK1 (Capsicum annuum ABA Induced MAP Kinase 1), corresponding to CaMEKK24/CA07g11550, plays a positive role in the modulation of ABA-dependent drought stress response.Citation11 To demonstrate this CaAIMK1 functional role in drought stress response, we generated CaAIMK1-silenced pepper plants using a tobacco rattle virus-based virus-induced gene silencing, and Arabidopsis transgenic plants that overexpress the CaAIMK1 driven by a 35S promoter.Citation11 These plants were subject to phenotypic, physiological, and biochemical analyses, including ABA-induced stomatal closing, leaf-surface temperature, transpiration water loss, expression of drought-inducible genes. Our data revealed that the silencing of CaAIMK1 in pepper plants led to reduced resistance to drought stress, while overexpression of CaAIMK1 in Arabidopsis plants enhanced drought resistance.Citation11 In this process, CaAIMK1 was involved in the regulation of leaf surface temperature and stomatal aperture in response to ABA. Also, the substitution of Lys-32 to Asn in the ATP-binding motif of CaAIMK1, leading to the loss of its kinase activity, did not promote drought resistance in Arabidopsis transgenic plants, indicating the kinase activity of CaAIMK protein was critical for drought resistance.Citation11
In conclusion, we found some of the pepper MEKK-like genes are potentially involved in ABA signaling and drought stress response and that CaAIMK1 functions as a positive kinase for drought resistance. Functional studies of the remaining genes are ongoing. The detailed mechanisms by which CaAIMK1 and other CaMEKK genes regulate drought resistance through ABA-dependent pathways and the members involved in the MAPK signaling cascade are still unknown. Identifying the CaAIMK1-interacting components in a MAPK cascade in pepper plants will help expand our understanding of the MAPK cascade in hormone signaling and stress response.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Xu J, Zhang SQ. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 2015;20:1–7. doi:10.1016/j.tplants.2014.10.001.
- Jagodzik P, Tajdel-Zielinska M, Ciesla A, Marczak M, Ludwikow A. Mitogen-activated protein kinase cascades in plant hormone signaling. Front Plant Sci. 2018:9:1387.
- de Zelicourt A, Colcombet J, Hirt H. The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 2016;21:677–685. doi:10.1016/j.tplants.2016.04.004.
- Meng XZ, Zhang SQ. MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol. 2013;51:245–266. doi:10.1146/annurev-phyto-082712-102314.
- Matsuoka D, Yasufuku T, Furuya T, Nanmori T. An abscisic acid inducible Arabidopsis MAPKKK, MAPKKK18 regulates leaf senescence via its kinase activity. Plant Mol Biol. 2015;87:565–575. doi:10.1007/s11103-015-0295-0.
- Colcombet J, Hirt H. Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J. 2008;413:217–226. doi:10.1042/BJ20080625.
- Mitula F, Tajdel M, CieślaA, Kasprowicz- MaluśkiA, Kulik A, Babula-SkowrońskaD, Michalak M, Dobrowolska G, Sadowski J, Ludwików A. Arabidopsis ABA-activated kinase MAPKKK18 is regulated by protein phosphatase 2C ABI1 and the ubiquitin-proteasome pathway. Plant Cell Physiol. 2015;56:2351–2367. doi:10.1093/pcp/pcv146.
- Danquah A, de ZélicourtA, Boudsocq M, Neubauer J, Frei Dit Frey N, Leonhardt N, Pateyron S, Gwinner F, Tamby J-P, Ortiz-Masia D, et al. Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J. 2015;82(2):232–244. doi:10.1111/tpj.12808.
- Danquah A, de Zelicourt A, Colcombet J, Hirt H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv. 2014;32:40–52. doi:10.1016/j.biotechadv.2013.09.006.
- Iftikhar H, Naveed N, Virk N, Bhatti MF, Song F. In silico analysis reveals widespread presence of three gene families, MAPK, MAPKK and MAPKKK, of the MAPK cascade from crop plants of Solanaceae in comparison to the distantly-related syntenic species from Rubiaceae, coffee. PeerJ. 2017;5:e3255. doi:10.7717/peerj.3255.
- Jeong S, Lim CW, Lee SC. The pepper MAP kinase CaAIMK1 positively regulates ABA and drought stress responses. Front Plant Sci. 2020;11:720. doi:10.3389/fpls.2020.00720.
- Colcombet J, Sozen C, Hirt H. Convergence of multiple MAP3Ks on MKK3 identifies a set of novel stress MAPK modules. Front Plant Sci. 2016;7:1941. doi:10.3389/fpls.2016.01941.
- Rodriguez MC, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol. 2010;61:621–649. doi:10.1146/annurev-arplant-042809-112252.
- Jonak C, Okresz L, Bogre L, Hirt H. Complexity, cross talk and integration of plant MAP kinase signalling. Curr Opin Plant Biol. 2002;5:415–424. doi:10.1016/S1369-5266(02)00285-6.
- Menges M, Doczi R, Okresz L, Morandini P, Mizzi L, Soloviev M, Murray JAH, Bgre L. Comprehensive gene expression atlas for the Arabidopsis MAP kinase signalling pathways. New Phytol. 2008;179:643–662. doi:10.1111/j.1469-8137.2008.02552.x.
- Wang RS, Pandey S, Li S, Gookin TE, Zhao ZX, Albert R, Assmann SM. Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. Bmc Genomics. 2011;12:216.
- Kong X, Pan J, Zhang D, Jiang S, Cai G, Wang L, Li D. Identification of mitogen-activated protein kinase kinase gene family and MKK-MAPK interaction network in maize. Biochem Biophys Res Commun. 2013;441:964–969. doi:10.1016/j.bbrc.2013.11.008.
- Wu J, Wang J, Pan C, Guan X, Wang Y, Liu S, He Y, Chen J, Chen L, Lu G. Genome-wide identification of MAPKK and MAPKKK gene families in tomato and transcriptional profiling analysis during development and stress response. PLoS One. 2014;9(7):e103032. doi:10.1371/journal.pone.0103032.
- Singh R, Lee MO, Lee JE, Choi J, Park JH, Kim EH, Yoo RH, Cho J-I, Jeon J-S, Rakwal R, et al. Rice mitogen-activated protein kinase interactome analysis using the yeast two-hybrid system. Plant Physiol. 2012;160:477–487. doi:10.1104/pp.112.200071.
- Liu Z, Shi L, Liu Y, Tang Q, Shen L, Yang S, Cai J, Yu H, Wang R, Wen J, et al. Genome-wide identification and transcriptional expression analysis of mitogen-activated protein kinase and mitogen-activated protein kinase kinase genes in Capsicum annuum. Front Plant Sci. 2015;6:780. doi:10.3389/fpls.2015.00780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi:10.1093/molbev/msy096.
- IchimuraK, Shinozaki K, Tena G, Sheen J, Henry Y, Champion A, Champion A, Kreis M, Zhang S, Hirt H, et al. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 2002;7:301–308. doi:10.1016/S1360-1385(02)02302-6.
- Li K, Yang F, Zhang G, Song S, Li Y, Ren D, Miao Y, Song C-P. AIK1, A mitogen-activated protein kinase, modulates abscisic acid responses through the MKK5-MPK6 kinase cascade. Plant Physiol. 2017;173:1391–1408. doi:10.1104/pp.16.01386.
- Li Y, Cai H, Liu P, Wang C, Gao H, Wu C, Yan K, Zhang S, Huang J, Zheng C. Arabidopsis MAPKKK18 positively regulates drought stress resistance via downstream MAPKK3. Biochem Biophys Res Commun. 2017;484(2):292–297. doi:10.1016/j.bbrc.2017.01.104.
- Tajdel M, Mitula F, Ludwikow A. Regulation of Arabidopsis MAPKKK18 by ABI1 and SnRK2, components of the ABA signaling pathway. Plant Signal Behav. 2016;11:e1139277. doi:10.1080/15592324.2016.1139277.
- Ludwikow A. Targeting proteins for proteasomal degradation-a new function of Arabidopsis ABI1 protein phosphatase 2C. Front Plant Sci. 2015;6:310.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi:10.1006/meth.2001.1262.
- Park C, Lim CW, Lee SC. The pepper CaOSR1 protein regulates the osmotic stress response via abscisic acid signaling. Front Plant Sci. 2016;7:890.
- Christmann A, Weiler EW, Steudle E, Grill E. A hydraulic signal in root-to-shoot signalling of water shortage. Plant J. 2007;52:167–174. doi:10.1111/j.1365-313X.2007.03234.x.
- Iuchi S, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol. 2000;123:553–562. doi:10.1104/pp.123.2.553.
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi:10.1093/nar/27.1.297.
