ABSTRACT
Phosphate (Pi) deficiency is one of the major adverse factors limiting plant growth and production. Enhanced RH development is thought to be the typical root morphological response under Pi deficiency, which will enhance the utilization of Pi resources from soil. Here, we report that MYB30-EIN3 module is functionally implicated in Pi deficiency-induced RH development in Arabidopsis. MYB30 and EIN3 antagonistically regulate RH growth via transcriptional regulation of RSL4 as well as other PSR genes, resulting in fine-tuned Pi uptake under Pi deficiency.
During their life cycle, plants are exposed to various environmental stresses. Among them, inorganic phosphate (Pi) deficiency is one of the major adverse factors limiting plant growth and production.Citation1 As an essential macronutrient, phosphorus serves as structural element of nucleic acids and cytomembrane, and participates in regulating multiple biological processes, including energy transfer, enzymatic activities, oxidative phosphorylation as well as signal transduction.Citation1,Citation2 Pi is distributed in soils unevenly, making a limited availability of it for plant growth and development.Citation1 Root hairs (RHs) are tubular outgrowths of the root epidermal cells, which account for a considerable area of root surface involved in nutrient and water uptake.Citation3 Enhanced RH development is thought to be the typical root morphological response under Pi deficiency, which will enhance the utilization of Pi resources from soil.Citation4 Several core genes implicated in the formation and development of RH have been identified, most of which encode specific transcription factors (TFs).Citation5–8 Functionally, these TFs form a cascade transcriptional network in regulating root epidermal cell fate specification and morphology.Citation5–7
The MYB proteins represent one of the largest plant TF family with a conserved N-terminal region required for DNA binding and a C-terminal modulator region responsible for their transcriptional activity.Citation9 The DNA-binding domain of MYB proteins might consist variable number of imperfect repeats (R) and based on the R number, the MYB TFs can be divided into different categories.Citation9 MYB30 belongs to the R2R3-MYB TF family, and it has been characterized to be function in plant growth and stress responses.Citation10,Citation11 Our previous study revealed that MYB30 negatively regulates the development of RH in Arabidopsis. We found that MYB30 could physically interact with ETHYLENE INSENSITIVE 3 (EIN3), a master TF in ethylene (ET) signaling, and they two antagonistically modulate the growth of RH via transcriptional regulation of RSL4 as well as other core RH genes.Citation8 Interestingly, it has been demonstrated that EIN3 is functionally involved in Pi deficiency-induced RH growth Citation12 and we therefore speculated that MYB30-EIN3 module might also function in the development of RH under Pi deficiency.
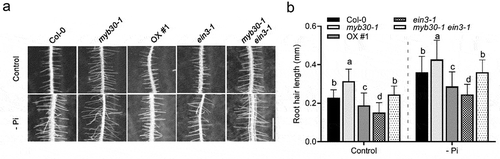
In the present study, we reported that MYB30 and EIN3 antagonistically regulate RH growth and phosphorus uptake under Pi deficiency condition in Arabidopsis. First, we determined whether MYB30 functions in RH growth under Pi deficiency.Citation4 Therefore, Col-0, myb30-1 and MYB30-OX plantsCitation8 were subject to RH development investigation under normal or Pi deficiency condition. Phenotypic data showed that RH growth was significantly enhanced in response to Pi deficiency in all the tested plants; however, the myb30-1 had longer RH whereas MYB30-OX transgenic plants had shorter RH compared to Col-0 in response to Pi deficiency, suggesting that MYB30 plays a negative role in regulating Pi deficiency-induced RH growth (). To further investigate the functional link between MYB30 and EIN3 in regulating RH growth under Pi deficiency, the myb30-1 ein3-1 mutant as well as Col-0 and myb30-1 plantsCitation8 were subject to RH phenotypic analysis under normal or Pi deficiency condition. Our data confirmed the positive role of EIN3 on RH elongation induced by Pi deficiency as the RH length in ein3-1 was shorter than that in Col-0 under Pi deficiency (). Importantly, we found that when grown under low Pi condition, the RH length in myb30-1 ein3-1 was shorter than myb30-1 but longer than ein3-1, suggesting that MYB30 and EIN3 antagonistically regulate RH development under Pi deficiency ().
Figure 1. MYB30 and EIN3 antagonistically modulate RH growth under the Pi deficiency condition. (a) RH length investigation. Seedlings of Col-0, myb30-1, MYB30-OX, ein3-1, and myb30-1 ein3-1 were germinated and grown on MS medium (Control) or MS medium without Pi (-Pi) for 4 d. Photographs were taken and the RH length was determined. Bar = 0.5 mm. (b) Quantification of the RH length in (a). Approximate 10 seedlings were scored for RH length determination in each experiment. Error bar represents SD (n > 10). Three independent experiments were carried out and similar results were obtained. Statistical significance was determined using a Student’s t-test by comparing with the WT control (Col-0) in each treatment; significant differences are indicated by different lowercase letters (P < .05)
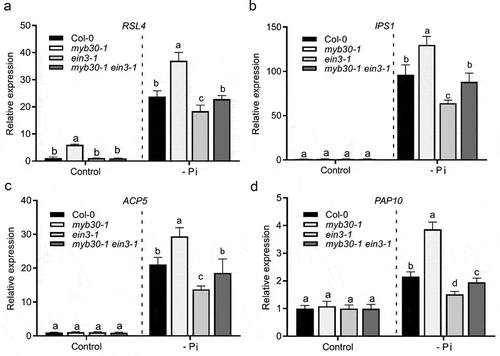
Our previous study has showed that RSL4 is a direct target of MYB30-EIN3 module, and that MYB30 and EIN3 antagonistically modulate RSL4 expression, leading to fine-tuned RH growth.Citation8 Next, we determined whether MYB30-EIN3 module is also implicated in the transcriptional regulation of RSL4 under Pi deficiency. RT-qPCR assays revealed that RSL4 was upregulated in myb30-1 plants but downregulated due to EIN3 deletion when Pi availability was limited (). Similarly, we found that several selected Pi starvation-responsive (PSR) genes inlcuding IPS1, ACP5 and PAP10 were upregulated in myb30-1 plants but downregulated in ein3-1 plants (–d). These results together demonstrated that MYB30 and EIN3 might antagonistically modulate expression of RSL4 and selected PSR-related genes. It is possible that MYB30-EIN3 module might serve as a switch of the transcriptional network for RH development, integrating developmental program and exogous stimulus (Pi deficiency).
Figure 2. MYB30 and EIN3 antagonistically modulate the expression of RSL4 and selected PSR genes under Pi deficiency. (a–d) RT-qPCR assays for the determination of the expression of RSL4 (a) and selected PSR genes (b-d). Seven-day-old Col-0, myb30-1 ein3-1 and myb30-1 ein3-1 plants were left untreated (Control) or grown under the -Pi condition. ACTIN2 was used as the internal control. Data were normalized by gene expression level in Col-0 under control condition. Error bar represents SD (n = 3). Statistical significance was determined using a Student’s t-test by comparing with the WT control (Col-0) in each treatment; significant differences are indicated by different lowercase letters (P < .05)
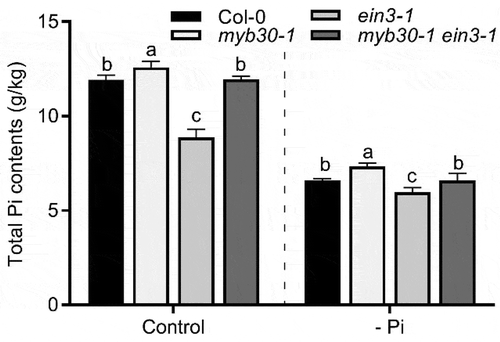
It has been found that the enhanced RH development is beneficial for plants to uptake Pi from soil.Citation13 Therefore, the total Pi content in the tested plants was determined as previously described.Citation14 The results showed that myb30-1 accumulated the highest level of Pi whereas ein3-1 plants accumulated the lowest level of Pi (). Interestingly, we found that the decreased Pi content in ein3-1 was partially rescued to a level like that of Col-0 in myb30-1 ein3-1 plants, indicating that MYB30 and EIN3 antagonistically modulate Pi uptake in plants ().
Figure 3. MYB30-EIN3 module regulates P uptake in Arabidopsis
Taken together, our data supported a conclusion that MYB30-EIN3 module is functionally implicated in Pi deficiency-induced RH development in Arabidopsis. MYB30 and EIN3 antagonistically regulate RH growth via transcriptional regulation of RSL4 as well as other PSR genes, resulting in fine-tuned P uptake under Pi deficiency. This study provides novel evidence for a better understanding of how plants respond to Pi deficiency, and provides essential candidate targets for engineering Pi deficiency-tolerant crops in the future.
Disclosure of potential conflicts of interest
The authors declare that there is no conflict of interests regarding the publication of this manuscript.
Acknowledgments
F.X. and Y.Z. conducted the experiments. Y.Z. and F.X. contributed to manuscript preparation with assistance of S.Z. H.Z. wrote the manuscript and supervised the project. All the authors contributed to discussion.
Additional information
Funding
References
- Nussaume L, Kanno S, Javot H, Marin E, Thibaud MC. Phosphate import in plants: focus on the PHT1 transporters. Front Plant Sci. 2011;2:1.
- Smith FW, Mudge SR, Rae AL, Glassop D. Phosphate transport in plants. Plant Soil. 2003;248:71–3.
- Rahman A, Hosokawa S, Oono Y, Amakawa T, Goto N, Tsurumi S. Auxin and ethylene response interactions during Arabidopsis root hair development dissected by auxin influx modulators. Plant Physiol. 2002;130:1908–1917.
- Song L, Yu H, Dong J, Che X, Dong L. The molecular mechanism of ethylene-mediated root hair development induced by phosphate starvation. PLoS Genet. 2016;12:e1006194.
- Grebe M. The patterning of epidermal hairs in Arabidopsis-updated. Curr Opin Plant Biol. 2012;15:31–37.
- Schiefelbein J, Huang L, Zheng X. Regulation of epidermal cell fate in Arabidopsis roots: the importance of multiple feedback loops. Front Plant Sci. 2014;5:47.
- Shibata M, Sugimoto K. A gene regulatory network for root hair development. J Plant Res. 2019;132:301–309.
- Xiao F, Gong Q, Zhao S, Lin H, Zhou H. MYB30 and ETHYLENE INSENSITIVE3 antagonistically modulate root hair growth in Arabidopsis. Plant J. 2021. doi:10.1111/tpj.15180.
- Liu J, Osbourn A, Ma P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant. 2015;8:689–708.
- Gong Q, Li S, Zheng Y, Duan H, Xiao F, Zhuang Y, He J, Wu G, Zhao S, Zhou H, et al. SUMOylation of MYB30 enhances salt tolerance by elevating alternative respiration via transcriptionally upregulating AOX1a in Arabidopsis. Plant J. 2020;102:1157–1171.
- Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylides C, Roby D. A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci USA. 2002;99:10179–10184.
- Zhang YJ, Lynch JP, Brown KM. Ethylene and phosphorus availability have interacting yet distinct effects on root hair development. J Exp Bot. 2003;54:2351–2361.
- Gahoonia TS, Nielsen NE. Root traits as tools for creating phosphorus efficient crop varieties. Plant Soil. 2004;260:47–57. doi:10.1023/B:PLSO.0000030168.53340.bc.
- Nagarajan VK, Jain A, Poling MD, Lewis AJ, Raghothama KG, Smith AP. Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol. 2011;156:1149–1163.
