ABSTRACT
The actin cytoskeleton plays pivotal roles in pollen tube growth by regulating organelle movement, cytoplasmic streaming, and vesicle trafficking. Previous studies have reported that plasma membrane-localized phospholipase Dδ (PLDδ) binds to cortical microtubules and negatively regulates plant stress tolerance. However, it remains unknown whether or how PLDδ regulates microfilament organization. In this study, we found that loss of PLDδ function led to a significant increase in pollen tube growth, whereas PLDδ overexpression resulted in pollen tube growth inhibition. We also found that wild-type PLDδ, rather than Arg 622-mutated PLDδ, complemented the pldδ phenotype in pollen tubes. In vitro biochemical assays demonstrated that PLDδ binds directly to F-actin, and immunofluorescence assays revealed that PLDδ in pollen tubes influences actin organization. Together, these results suggest that PLDδ participates in the development of pollen tube growth by organizing actin filaments.
Introduction
In flowering plants, double fertilization within the pistil is achieved through growth of the tip of the pollen tube. Many pharmacological and genetic studies have indicated that the actin filament is a major mediator of pollen tube tip growth in plants.Citation1–5 Polar pollen tube growth is regulated by several cellular and extracellular components including reactive oxygen species and the cytoskeleton, as well as processes such as vesicular traffickingCitation6–8 for targeted synthesis of new plasma membrane and cell wall components.Citation9
The plasma membrane acts as both a biological barrier and a location for the sensation of extracellular stimuli. Phospholipids are the main component of the plasma membrane, and phospholipase D (PLD), which hydrolyzes phospholipids, is involved in important physiological and biochemical processes such as signal transduction, membrane transport, cytoskeletal rearrangement, and membrane degradation.Citation10–14 The PLD gene family of Arabidopsis thaliana encodes 12 functional proteins, which are classified into six types: PLDα (3), β (2), γ (3), δ, ε, and ζ (2).Citation15 Among these, PLDδ exhibits high expression in the pollen tube.Citation16 PLDδ physically interacts with both the microtubule and actin cytoskeleton.Citation13 Studies of tobacco have shown that PLDβ1 activity can be increased by F-actin and inhibited by G-actin.Citation17 In Arabidopsis, plasma membrane-localized PLDδ has been found to negatively regulate plant heat tolerance by depolymerizing cortical microtubules.Citation18 Recent studies have suggested that self-incompatibility-induced phospholipid acid (PA) derived from PLDδ in pollen delays actin cytoskeleton depolymerization.Citation19 Moreover, overexpression of tobacco PLDδ causes significant reduction in pollen tube growth.Citation20 These findings indicate that PLDδ is tightly associated with cytoskeletal organization; therefore, in the present study, we examined the role of PLDδ in mediating pollen tube growth via the actin cytoskeleton.
Results and discussion
Given that Arabidopsis PLDδ is highly expressed in the pollen tube, it is reasonable to speculate that PLDδ is involved in pollen tube growth regulation. In this study, we used two homozygous T-DNA insertion lines, termed pldδ-1 (SALK_092469) and pldδ-2 (SALK_023247), to assess pollen tube behavior in vitro.Citation18 Both mutants exhibited similar pollen germination rates to wild-type (WT), but with longer pollen tubes (). Analysis of these results showed that pollen tubes containing pldδ were significantly longer than those of WT plants (), and pollen tube growth rates of pldδ-1 were higher than those in WT ().
Figure 1. PLDδ interacts with actin filaments and regulates pollen tube growth in Arabidopsis.
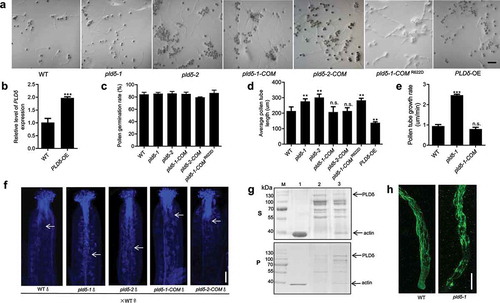
To determine whether pollen tube defects were caused by loss of PLDδ function, we performed a complementation experiment. The introduction of PLDδ into pldδ-1 and pldδ-2 (pldδ-1-COM and pldδ-2-COM) completely rescued the pldδ phenotype, whereas the Arg (R) 622 mutation of PLDδ, which resulted in almost complete loss of PLDδ activity,Citation18,Citation21 failed to restore the phenotypes of pollen tube growth (). This result demonstrates that Arg (R) 622 may be the crucial site for PLDδ regulation of pollen tube growth. We also generated a PLDδ-overexpressing line under the control of pollen-specific promoter Lat52. Reverse transcription quantitative PCR (RT-qPCR) analyses showed that PLDδ transcript level was increased approximately twofold in pollen compared to WT (). Pollen tube elongation was sharply inhibited in the overexpression line (). These results demonstrate that PLDδ negatively regulates pollen tube growth.
To directly observe pollen tube behavior, we pollinated WT stigmas with pollen derived from WT, pldδ-mutant, and PLDδ-complementation lines. At 8 h after pollination, pollinated pistils were fixed and stained with aniline blue and observed by confocal microscopy. We found that pollen tubes of pldδ mutants had penetrated through the style and reached the top of the transmitting tract (). By contrast, pollen tubes from WT and both PLDδ-complemented lines had only begun to penetrate the style, suggesting that pollen tube growth in the style and transmitting tract is accelerated in pldδ mutants.
The actin cytoskeleton is crucial for pollen tube growth;Citation4,Citation22–24 it acts as a structural element that supports the turgor pressure required to drive and maintain rapid pollen tube growth. PLD-derived PA interacts directly with capping proteins, leading to their separation from actin, which promotes actin polymerization.Citation25,Citation26 PA also binds to MAP65-1 and mediates microtubule stabilization,Citation27 and plasma membrane-associated PLDδ negatively regulates plant thermotolerance via destabilizing cortical microtubules.Citation18 Moreover, Arabidopsis PLDδ decorated subapical plasma membrane in tobacco pollen tube, but no plasma membrane signal was found at the tip.Citation20 Considering that PLDδ might also function as an actin-binding protein in pollen tubes, we performed a co-sedimentation assay to investigate whether PLDδ binds directly to F-actin. The resulting supernatants and pellets were analyzed separately by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE). The results showed that PLDδ co-sedimented with F-actin and that higher levels of PLDδ were present in pellets containing F-actin than in those without F-actin (); these results are consistent with those of a previous study that reported PLDδ interaction with actin based on epitope-tagged affinity pull-down assays.Citation13
Actin cytoskeleton drives the intracellular transport system, and the dynamic organization of actin cytoskeleton is essential for pollen tube growth.Citation2,Citation28,Citation29 We next observed F-actin organization in pollen tubes using immunostaining under a confocal microscope. In WT plants, the growing pollen tubes produced large amounts of actin cables that became disorganized near the tip region (). By contrast, clear disorganization of the microfilament bundles was apparent in the pldδ-1 mutant, which formed fragmented and less axially oriented microfilaments (), indicating that PLD may regulate the organization of actin filaments. It should be noted that both excess stabilization and destabilization of actin filaments will lead to retarded pollen tube growth.Citation2,Citation28 Together, our findings suggest that PLDδ regulates F-actin dynamics in pollen tubes; therefore, we propose a phospholipid-based mechanism underlying pollen tube growth regulation. Certainly, the potential role for PLDδ and its detailed mechanisms in regulation of the turnover of actin filaments in pollen tube needs to be examined in the future.
Methods
Plant materials and pollen germination conditions
In this study, we used the Arabidopsis (Arabidopsis thaliana) ecotype Columbia. The identification and verification of the T-DNA-insertion mutant and complement lines of PLDδ were reported previously.Citation18 Arabidopsis pollen was isolated from newly opened flowers and placed on pollen germination medium [1 mM CaCl2, 1 mM Ca(NO3)2, 1 mM MgSO4, 0.01% (w/v) H3BO3, and 18% (w/v) sucrose solidified with 0.8% (w/v) agar, pH 7.0]. The plates were cultured at 27°C under moist conditions. Pollen germination and pollen tubes were observed using a Zeiss fluorescence microscope equipped with differential interference contrast optics. The pollen germination percentage and average pollen tube length were calculated based on at least three experiments, each measuring 50 pollen tubes. The ImageJ software (National Institutes of Health) was used to measure pollen tube length.
Plasmid construction
To generate the PLDδ overexpression line, PLDδ was cloned from Arabidopsis cDNA by polymerase chain reaction (PCR) and subcloned into the pSUPER 1300 vector between the site of SalI and SpeI, driven by the pollen-specific promoter Lat52 for transformation into Arabidopsis. Primers used for PCR amplification are as follows: 5ʹ-CGCGGATCCATACTCGACTCAGAAGGTATTGAGG-3ʹ (BamHI) and 5ʹ-AAAGTCGACTAATTGGAAATTTTTTTTTTGGTGT-3ʹ (SalI) for Lat52 promoter; 5ʹ-ACGCGTCGACATGGCGGAGAAAGTATCGGAGGA-3ʹ (SalI) and 5ʹ-CGGACTAGTTTACGTGGTTAAAGTGTCA GGAAGA-3ʹ (BcuI) for PLDδ.
RNA extraction and RT-qPCR
Total RNA from pollen grains were collected from open flowers and extracted using RNAiso regent (Takara). PLDδ expression level was analyzed by reverse transcription quantitative PCR (RT-qPCR) using SYBR mix. The primers used for PLDδ were 5ʹ- ACAACAGGCAAGTCCTAGCTC-3ʹ and 5ʹ-GCAAATCGTGCCAAGGTTGT −3ʹ. UBQ5 was used as internal control. The primers used for UBQ5 were 5ʹ- CGTGAAAACCCTAACGGGGA −3ʹ and 5ʹ- GACTCGCCATGAAAGTCCCA −3ʹ.
Aniline blue staining assay
Pre-emasculated mature WT flowers were pollinated with pollen from WT, pldδ-1, pldδ-2, pldδ-1-COM, and pldδ-2-COM. After 8 h, the pollinated pistils were fixed in fixing solution containing ethanol:acetic acid (3:1) for 2 h at room temperature and then washed with distilled water three times (5 min each); the pistils were then incubated in 8 M NaOH solution overnight. The following day, the pistils were washed in distilled water three times (1 h each). The pistils were then stained in aniline blue solution (0.1% aniline blue in 0.1 M K2HPO4-KOH buffer, pH 11.0) for 5 h in the dark and then were observed under the confocal laser scanning microscope (Zeiss 780). The ultraviolet excitation wavelength maximum was 350 nm, and the emission spectra were collected from 440 to 612 nm.
Protein purification and co-sedimentation assay
PLDδ cDNA was amplified from plant total cDNA and linked to the PGEX-4 T-1 vector. The plasmid was transformed into Escherichia coli (BL21). Protein expression was induced at 23°C overnight with 1 mM isopropyl-β-d-1-thiogalactopyranoside. To purify the protein, the harvested bacterial cells were resuspended in phosphate-buffered saline (PBS) containing 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4, with 1 mM phenylmethylsulfonyl fluoride and 1 mg/mL lysozyme. After incubation on ice for 30 min, samples were subjected to ultrasonic treatment and centrifuged at 10,000 × g for 30 min at 4°C. GST-PLDδ was then affinity purified using glutathione resin (GenScript, Nanjing, China) and subjected to SDS-PAGE.
For the co-sedimentation assays, we prepared 700 μL F-actin buffer (630 μL general actin buffer and 70 μL actin polymerization buffer) in a centrifuge tube on ice. Actin was incubated with GST-PLDδ (20 μg) in the F-actin buffer for 30 min. After centrifugation at 40,000 g for 20 min at 25°C, the resulting supernatants and pellets were separated. The pellets and supernatants were combined with Laemmli reducing sample buffer (4% [w/v] SDS, 20% [v/v] glycerol, 125 mm Tris-HCl, pH 6.8, 0.004% [v/v] bromophenol blue) and then analyzed by SDS-PAGE using 10% acrylamide gels.
Immunofluorescence staining
The pollen tubes were stained after growth on solid germination medium at 27°C for 2 h. Pollen tubes were fixed in fixative solution (4% paraformaldehyde, 1 mM MgCl2, 1 mM EGTA, 10% sucrose, and 100 mM PIPES buffer, pH 6.9) for 1.5 h and then washed gently with PBS buffer three times. The fixed pollen tubes were treated with digestion buffer (1% cellulose and 0.5% pectinase in PBS buffer) at room temperature for 45 min and then washed gently with PBS containing 0.1% Nonidet P-40 three times. The pollen tubes were incubated in primary antibody at 4°C overnight and then incubated in secondary antibody at 37°C in germination medium for 2 h. The primary antibody was diluted to 1:600 in liquid germination medium containing 5% Gly with mouse anti-F-actin monoclonal antibody (CMCTAG, China). Alexa 488-conjugated goat anti-mouse IgG (diluted to 1:400, Invitrogen) was used as the secondary antibody.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Xiang Y, Huang X, Wang T, Zhang Y, Liu Q, Hussey PJ, Ren H. ACTIN BINDING PROTEIN29 from lilium pollen plays an important role in dynamic actin remodeling. Plant Cell. 2007;19:1–4. doi:10.1105/tpc.106.048413.
- Wu Y, Yan J, Zhang R, Qu X, Ren S, Chen N, Huang S. Arabidopsis FIMBRIN5, an actin bundling factor, is required for pollen germination and pollen tube growth. Plant Cell. 2010;22(11):3745–3763. doi:10.1105/tpc.110.080283.
- Su H, Zhu J, Cai C, Pei W, Wang J, Dong H, Ren H. FIMBRIN1 is involved in lily pollen tube growth by stabilizing the actin fringe. Plant Cell. 2012;24:4539. doi:10.1105/tpc.112.099358.
- Gibbon BC, Kovar DR, Staiger CJ. Latrunculin B has different effects on pollen germination and tube growth. Plant Cell. 1999;11:2349–2363. doi:10.1105/tpc.11.12.2349.
- Chen CY, Wong EI, Vidali L, Estavillo A, Hepler PK, Wu HM, Cheung AY. The regulation of actin organization by actin-depolymerizing factor in elongating pollen tubes. Plant Cell. 2002;14:2175–2190. doi:10.1105/tpc.003038.
- Samaj J, Müller J, Beck M, Böhm N, Menzel D. Vesicular trafficking, cytoskeleton and signalling in root hairs and pollen tubes. Trends Plant Sci. 2006;11:594–600. doi:10.1016/j.tplants.2006.10.002.
- Cárdenas L. New findings in the mechanisms regulating polar growth in root hair cells. Plant Signal Behav. 2009;4:4–8. doi:10.4161/psb.4.1.7341.
- Guan Y, Guo J, Li H, Yang Z. Signaling in pollen tube growth: crosstalk, feedback, and missing links. Mol Plant. 2013;6:1053–1064. doi:10.1093/mp/sst070.
- Barberini ML, Muschietti J. Imaging of calcium dynamics in pollen tube cytoplasm. Methods Mol Biol. 2015;1242:49–57.
- Qin C, Wang X. The Arabidopsis phospholipase D family. Characterization of a calcium-independent and phosphatidylcholine-selective PLD zeta 1 with distinct regulatory domains. Plant Physiol. 2002;128:1057–1068. doi:10.1104/pp.010928.
- Hong Y, Devaiah SP, Bahn SC, Thamasandra BN, Li M, Welti R, Wang X. Phospholipase D epsilon and phosphatidic acid enhance Arabidopsis nitrogen signaling and growth. Plant J. 2009;58:376–387. doi:10.1111/j.1365-313X.2009.03788.x.
- Lin DL, Yao HY, Jia LH, Tan JF, Xu ZH, Zheng WM, Xue H-W. Phospholipase D-derived phosphatidic acid promotes root hair development under phosphorus deficiency by suppressing vacuolar degradation of PIN-FORMED2. New Phytol. 2020;226:142–155. doi:10.1111/nph.16330.
- Ho AYY, Day DA, Brown MH, Marc J. Arabidopsis phospholipase Dδ as an initiator of cytoskeleton-mediated signalling to fundamental cellular processes. Fun Plant Biol. 2009;36:190–198. doi:10.1071/FP08222.
- Motes CM, Pechter P, Yoo CM, Wang YS, Chapman KD, Blancaflor EB. Differential effects of two phospholipase D inhibitors, 1-butanol and N-acylethanolamine, on in vivo cytoskeletal organization and Arabidopsis seedling growth. Protoplasma. 2005;226:109–123. doi:10.1007/s00709-005-0124-4.
- Wang X. Regulatory functions of phospholipase D and phosphatidic acid in plant growth, development, and stress responses. Plant Physiol. 2005;139:566–573. doi:10.1104/pp.105.068809.
- Zhang WWX, Hong Y, Li W, Wang X. Plant phospholipase D. Lipid Signaling Plants. 2010;23:39–60.
- Pleskot R, Potocký M, Pejchar P, Linek J, Bezvoda R, Martinec J, Valentová O, Novotná Z, Žárský V. Mutual regulation of plant phospholipase D and the actin cytoskeleton. Plant J. 2010;62:494–507. doi:10.1111/j.1365-313X.2010.04168.x.
- Zhang Q, Song P, Qu Y, Wang P, Jia Q, Guo L, Zhang C, Mao T, Yuan M, Wang X, et al. Phospholipase Dδ negatively regulates plant thermotolerance by destabilizing cortical microtubules in Arabidopsis. Plant Cell Environ. 2017;40:2220–2235. doi:10.1111/pce.13023.
- Chen J, Wang P, De Graaf BHJ, Zhang H, Jiao H, Tang C, Zhang S, Wu J. Phosphatidic acid counteracts S-RNase signaling in pollen by stabilizing the actin cytoskeleton. Plant Cell. 2018;30(5):1023–1039. doi:10.1105/tpc.18.00021.
- Pejchar P, Sekeres J, Novotny O, Zarsky V, Potocky M. Functional analysis of phospholipase Delta family in tobacco pollen tubes. Plant J. 2020;103:212–226. doi:10.1111/tpj.14720.
- Song P, Jia Q, Chen L, Jin X, Xiao X, Li L, Chen H, Qu Y, Su Y, Zhang W, et al. Involvement of Arabidopsis phospholipase D delta in regulation of ROS-mediated microtubule organization and stomatal movement upon heat shock. J Exp Bot. 2020;71:6555–6570. doi:10.1093/jxb/eraa359.
- Fu Y. The actin cytoskeleton and signaling network during pollen tube tip growth. J Integr Plant Biol. 2010;52:131–137. doi:10.1111/j.1744-7909.2010.00922.x.
- Zhu J, Nan Q, Qin T, Qian D, Mao T, Yuan S, Wu X, Niu Y, Bai Q, An L, et al. Higher-ordered actin structures remodeled by Arabidopsis ACTIN-DEPOLYMERIZING FACTOR5 are important for pollen germination and pollen tube growth. Mol Plant. 2017;10:1065–1081. doi:10.1016/j.molp.2017.06.001.
- Li G, Yang X, Zhang X, Song Y, Liang W, Zhang D. Rice morphology determinant-mediated actin filament organization contributes to pollen tube growth. Plant Physiol. 2018;177:255–270. doi:10.1104/pp.17.01759.
- Huang S, Blanchoin L, Kovar DR, Staiger CJ. Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. J Biol Chem. 2003;278:44832–44842. doi:10.1074/jbc.M306670200.
- Huang S, Gao L, Blanchoin L, Staiger CJ. Heterodimeric capping protein from Arabidopsis is regulated by phosphatidic acid. Mol Biol Cell. 2006;17:1946–1958. doi:10.1091/mbc.e05-09-0840.
- Zhang Q, Lin F, Mao T, Nie J, Yan M, Yuan M, Zhang W. Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. Plant Cell. 2012;24:4555–4576. doi:10.1105/tpc.112.104182.
- Jiang Y, Wang J, Xie Y, Chen N, Huang S. ADF10 shapes the overall organization of apical actin filaments by promoting their turnover and ordering in pollen tubes. J Cell Sci. 2017;130:3988–4001. doi:10.1242/jcs.207738.
- Qu X, Jiang Y, Chang M, Liu X, Zhang R, Huang S. Organization and regulation of the actin cytoskeleton in the pollen tube. Front Plant Sci. 2014;5:786.
