ABSTRACT
American ginseng, a valuable medicinal and food plant, is threatened by rot root, which affects its yield and quality. However, limited studies have investigated the changes in soil microbial community and physiochemical properties between healthy and rot root American ginseng. Here, high-throughput sequencing and soil physiochemical properties were used to characterize these changes. The soil physiochemical properties showed significance differences between the soil of healthy and rot root, in which the pH, available potassium, available phosphorus, soil organic carbon and soil organic matter were significantly higher in healthy root soil. Besides, fungal α-diversity was also higher in healthy root soil than that in rot root. Importantly, the dominant fungal genera differed between soils of healthy and rot root of American ginseng, and LEfSe further indicated that six fungal genera (Devriesia, Chrysosporium, Dichotomopilus, Pseudeurotium, Acaulium and Scedosporium) were significantly enriched in the soil of healthy plants, whereas six fungal genera (Gibellulopsis, Fusarium, Plectosphaerella, Tetracladium, Gibberella and Ilyonectri) were significantly enriched in the soil of rot root, suggesting that an increase in the relative abundance of these pathogenic fungi (Fusarium, Plectosphaerella, and Ilyonectri) may be associated with ginseng rot root. Notably, this study is the first to report that an increase in the relative abundances of Gibellulopsis and Gibberella in the rot root soil of American ginseng may be associated with the onset of rot root symptoms in this plant. The functional profile prediction showed that the there was a significantly Pathotrophs increase in the rot root soil compared with healthy root soil and Saprotrophs were more abundant in the healthy root soil. Finally, correlation analyses revealed that soil cation exchange capacity was an important factors affecting the composition of rot root of American ginseng soil microbial communities. This study not only used a new approach to explore the new fungal associated with rot root in American ginseng but also excavated the major soil physiochemical properties affecting the microbiome diversity, providing foundation for developing biocontrol strategies against rot root.
Introduction
American ginseng (Panax quinquefolius L.) is a perennial herb of Panax genus.Citation1 Native to the moist, deciduous forests of eastern North America, it is now grown in China, Japan, Korea and Russia.Citation2,Citation3 Ginsenoside is the primary chemical component of American ginseng.Citation4 Modern pharmacological studies have shown that it has a wide range of therapeutic properties, including hypoglycemic,Citation5–8 immunomodulatoryCitation9–13 and neurotrophic effects,Citation14,Citation15 and anticancer activities.Citation16,Citation17 Considering the high medicinal and economic value of American ginseng, it has been listed as the homology product of medicine and food by the National Health Commission and the State Administration for Market Regulation in 2020.Citation18
American ginseng is susceptible to infection with fungal pathogens because of its special growing conditions.Citation19 Seeds germinate for two years, and plants usually grow for more than four years before being harvested.Citation20 During the growth process of American ginseng, it requires low temperatures, low light and moist soil, which are ideal growth conditions for fungal pathogens contributing to the plant disease. Rot root, characterized by reddish-brown to orange-brown discoloration areas on the root surface, is one of the most destructive plant diseases affecting American ginseng quality.Citation21 The fungal identification rot root in American ginseng is primarily based on the isolation and culture of fungi at present. The morphological and microscopic characteristics of pure isolates are usually analyzed in combination with DNA sequences.Citation22,Citation23 However, the isolation and culture of fungi is a complex and time-consuming process, with the risk of losing some strains, resulting in inaccurate characterizations of fungal diversity.
High-throughput sequencing (HTS) is a growth-independent approach that can provide large amounts of data on the composition of low-abundance mixed microbial communities, such as soil, sediment and air filter samples.Citation24 It has been employed to discovery of vital microbiomes associated with plant disease.Citation25 The study of soil microbial communities between rusty and healthy ginseng root showed that there was an increase in pathogenic microorganisms such as Ilyonectria and a reduction of beneficial microorganisms such as Tremellomycetes Acidobacteria subgroup 6 and Gemmatimonadetes in Ginseng rusty root.Citation26Moreover, rot root affected the community structure and diversity of endophytic fungi in the rhizosphere and root of Panax notoginseng.Further, the occurrence of plant disease is related to the deterioration of soil physical and chemical properties.Citation28 However, limited studies are exploring the comprehensive microbiomes associated with the rot root of American ginseng and the relationship between soil microbiomes and physicochemical properties of rot root of American ginseng.
In Citation27 this study, we compared the soil physicochemical properties and microbial communities in of both healthy and rot root American ginseng. Moreover, the relationship of soil physicochemical properties and microbial communities was explored. Finally, the functional profile of healthy and rot root American ginseng was also studied. The research results will help to understand the contribution of soil microecology to rot root etiology and promote the formulation of effective biological control strategies and the sustainable development of traditional medicine industry.
2. Material and methods
2.1. Sample collection
In October 2020, we collected the rot and healthy root and its corresponding soil samples in Huoshaodian Township, Liuba County, Shannxi provinces, one of the major producing areas of American ginseng in China (Table 1). Voucher specimens with voucher numbers from 20201013019-PQ to 20201013030-PQ were deposited in the herbarium of the Institute of Medicinal Plant Development at the Chinese Academy of Medical Sciences in Beijing, China, which was identified as Panax quinquefolium by professor Linfang Huang. Six healthy roots (HS:HS1,HS2,HS3,HS4,HS5 and HS6)soil and six rot roots (RS:RS1,RS2,RS3,RS4,RS5 and RS6) soil of American ginseng were collected in different places in Huoshaodian Township. Soil cores were taken at a depth of 5 cm using a stainless-steel cylindrical driller with a diameter of 5 cm and then in liquid nitrogen. After being transported to the laboratory, the soil samples were passed through a 2 mm sieve to remove plant tissues, roots, rocks, and other debris and then stored at −80°C in a refrigerator before further experiments.
2.2. Soil physicochemical properties
Physicochemical parameters including soil texture, moisture content, pH, available phosphorus, available potassium, ammonium N, organic matter (SOM), cation exchange capacity(CEC) and soil organic carbon (SOC) were determined by standard methods (LY/T 1225–1999, NY/T52-1987, LY/T1239-1999, LY/T 1232–2015, LY/T 1234–2015 and LY/T1228-2015, respectively,). Each experiment was repeated in triplicate.
2.3. DNA extraction and sequencing
The microbial DNA of 12 soil samples (six healthy roots and six rot roots) of American ginseng was extracted using the HiPure soil DNA Kits ((Magen, Guangzhou, China) according to manufacturer’s protocols. For each sample, DNA was used to amplify the fungal ITS2 region ITS3/KYO2 and ITS4).Citation29 PCR reactions were performed in triplicate 50 uI mixture containing 5 uL of 10× KOD Buffer,5 uL of 2 mM dNTPs,3 LL of25mM MgSO4,1.5 uL of each primer(10 uM),1 uL of KOD Polymerase, and 100 ng of template DNA. Amplicons were extracted from 2% agarose gels and purified using the AxyPrep DNA GelExtraction Kit(Axygen Biosciences, Union City, CA,U.S.) according to the manufacturer’s instructions and quantified using ABI StepOnePlus Real-Time PCR System(Life Technologies, Foster City, USA). Purified amplicons were pooled in equimolar and paired-end sequenced (PE250) on an Illumina platform according to the standard protocols.
To get high quality clean reads, raw reads were further filtered according to the specification. After clean reads were merged as raw tags, raw tags were filtered according to the standard pipeline to obtain the high-quality clean tags. The clean tags were clustered into operational taxonomic units (OTUs) of ≥ 97% similarity. Finally obtained effective tags for further analysis. The tag sequence with highest abundance was selected as representative sequence within each cluster.
2.4. Bioinformatics and statistical analysis
The representative OTU sequences were classified into organisms by a naive Bayesian model ITS2 database (version update_2015),Citation30 with the confidence threshold value of 0.8. The circular layout representations of species abundance were graphed using CIRCOS (version 0.69–3).Citation31
Alpha diversity was employed to analyze the complexity of species diversity using five indexes, namely, Chao1, observed species and ACE. These three indices were calculated using the QIIME software.Citation32 Alpha index comparison between groups was calculated by Wilcoxon rank test in R project Vegan package. Beta diversity analysis was used to evaluate differences of samples in terms of species complexity. Beta diversity was calculated using the principal coordinate analysis(PCoA). PCoA of Unweighted unifrac was generated in R project Vegan package (version 2.5.3) and plotted in R project ggplot2 package (version 2.2.1).Citation33
Biomarker features in each group were screened by LEfSe softwareCitation34(version 1.0)(LDA>3.5 and p < .05) and randomforest package [25] (version 4.6.12) in R project. For functional profile, the FUNGuild v1.0 database was used to assign ecological functions (trophic modes) to each OTU.Citation35
2.5. Correlation analysis of relative high abundance microbiomes and soil physicochemical properties
Redundancy analysis(RDA) of relative high abundance microbiomes and soil physicochemical properties was performed using Canoco 5 software. Principal components analysis (PCA) was first used to reduce the number of environmental variables taken forward to further analyses. Variables that significantly explained variation in ginsenosides were determined with forward selection (999 Monte Carlo permutations; false discovery rate (FDR) p < .05) and used in RDA.
3. Results
3.1 Soil physicochemical properties
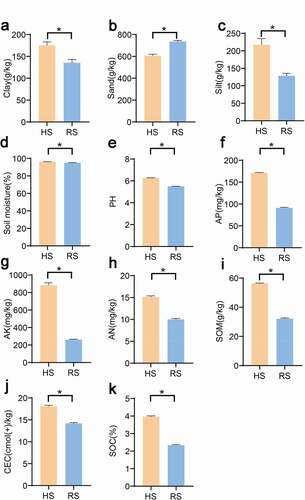
The major physicochemical properties of the soil samples are presented in . The particle distribution analysis of the soil samples reveals that all soil samples were sandy loam. The soil samples tend toward acidic with pH of 5.50 for RS, while HS soil has a higher pH of 6.26. In compared with HS soil, RS soil appeared nutrition deficiency (defined by low of detectable available potassium, available phosphorus, ammonium N, SOC and SOM. This indicated that the soil physicochemical properties of the HS and RS soil is significantly different.
Figure 1. Soil physiochemical characteristics between RS and HS soil of American ginseng. a: clay; b: sand; c: silt; d: soil moisture content; e: pH; f: available phosphorus; g: available potassium; h: Ammonium N; i: Soil organic carbon; j: Soil organic matter; k: Cation Exchange Capacity;. “*”represent significant differences, p < .01. HS: healthy root; RS: rot root
3.2. Microbiomes composition
The clean reads were obtained in the 12 samples, and the number varied from 120,610 to 136,619 (Table 1). These sequences were divided into 1,107 OTUs after cluster analysis. The number of unique and common OTUs for the two groups are shown in a Venn diagram(Figure S1). The results showed that the HS group possessed more unique OTUs than the RS group. Of the OTUs, 376 and 259 were respectively unique for HS and RS soil groups, and the remaining 224 were shared by both of the groups.
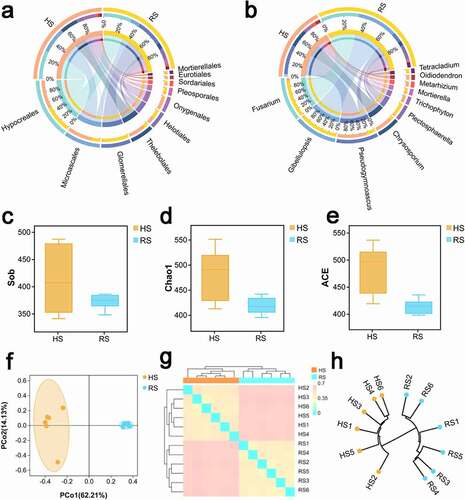
The microbiomes composition of HS and RS soil differed. The fungal community was identified into 12 phyla, 72 orders and 266 genera. The circos plot generated by using CIRCOS software, visualizes the similarities and differences between HS and RS soil microbiomes. At the order level(), Hypocreales tend to be the dominant between the two groups (HS,27.37%;RS, 27.18%). Following that, Microascales (23.75%) tilted toward HS soil, unlike Glomerellales (25.38%) that was abundant in RS. At the genus level (), Pseudogymnoascus (14.03%) and Chrysosporium (8.24%) were dominant in HS soil, whilst Gibellulopsis (19.00%) and Fusarium (18.41%) were abundant in RS. This revealed that the composition of HS and RS soil differed.
Figure 2. The microbial community composition and diversity in HS and RS soil of American ginseng (a) Distribution of bacterial community for each group at the order level were visualized by Circos. (b) Distribution of bacterial community for each group at the genus level were visualized by Circos. The upper half circle indicates the species composition in each group: the color of the outer ribbon represents different groups; the color of the inner ribbon represents the composition of different species in group, and the length of the ribbon represents the relative abundance of the corresponding species. The lower half circle indicates the distribution ratio of species in different group at the genus level: the outer ribbon represents the genus; the inner color of the ribbon represents different groups, and the length represents the proportion of the sample for a particular genus.α-diversity:(c)Sob;(d)Chao1;(e)ACE.β-diversity:(f)PCoA plot based on the unweighted UniFrac distance matrix.(g) Heat map on the basis of Bray-Curtis distance matrix. (h) Cluster Dendrogram based on Jaccard distance. HS: healthy root; RS: rot root
3.3. Microbiomes diversity
The measurements of α-diversity revealed that the diversity of RS soil decreased compared with HS soil. The α-diversity of soil fungal communities in each sample was evaluated on the observed species diversity(),Chao 1() and ACE(). The Chao 1, observed species diversity and ACE indexes suggested that α-diversity was higher than that in the HS soil compared with the RS soil. The results of good’s coverage which is an index of sampling completeness, indicated good overall sampling with levels of >97%(Figure S2a). Rarefaction curve analysis showed that all samples were almost parallel to the x-axis, thereby indicating that the obtained reads were sufficient to represent the overall fungal diversity (Figure S 2b). For β-diversity, The results of the unconstrained principal coordinate analysis (PCoA) of unweighted UniFrac distance 2D plots indicated that the soil samples of fungal () in different kind soil were well clustered. The PCoA PC1 and PC2 represented 62.21% and 14.13%, respectively, of the variance, and the contribution of the cumulative variance of the two principal coordinates (PC1 and PC2) accounted for 76.34%. Anosim results (Figure S3) also showed that there are significant differences (p < .01) among the HS and RS soils. Heat maps on the basis of the Bray-Curtis distance matrix (g) and Cluster Dendrogram based on Jaccard distance () of the soil samples demonstrated that the HS soil were closely clustered and RS soil were also closely clustered.
3.4. Biomarker microbiomes determination
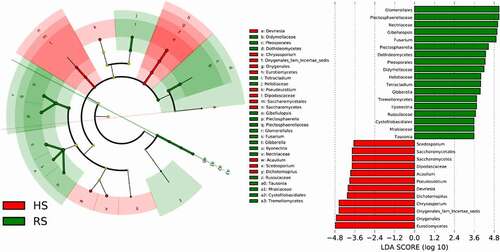
displays that the fungal biomarker microbiomes from the phylum to genus level between the rot root soil and healthy root soil samples were different. The LEfSe methods were used to identify features with significant differential abundance between the soil samples, calculate the effect size of each differentially abundant features and determine the biomarker fungal microbiome in the two different soil at the genus level. The results presented in Table S1 revealed that at the class level, the LDA score of GS35, Saccharomycetes, Eurotiomycetes and Pezizomycetes class were much higher in HS soil samples, whereas the RS soil samples feature higher number of Tremellomycetes and Dothideomycetes. At the genus level, the LDA score of Devriesia, Chrysosporium, Dichotomopilus, Pseudeurotium, Acaulium and Scedosporium was the highest in the HS soil microbiomes, whilst that of Gibellulopsis, Fusarium, Plectosphaerella, Tetracladium, Gibberella and Ilyonectri was the highest in RS soil.
Figure 3. Differential microbial profiles of American ginseng HS and RS soil. Linear discriminant analysis effect size(LEfSe) for microbial communities from phyla to genera in rot and health root soils. Green circles represent taxa that were significantly abundant in RS soil, while red circles represent taxa that were significantly abundant in HS soil. Only taxa meeting a linear discriminant analysis significance threshold of > 3.5 and p ≤ 0.05 were shown and color-coded. HS: healthy root; RS: rot root
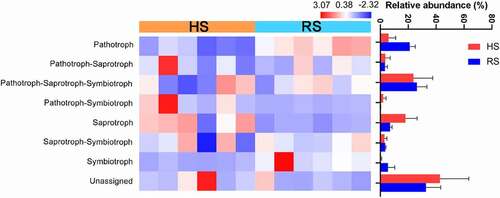
3.5. Prediction of the microbial functional profiles of the microbiome
We used FUNGuild to assign functional roles to OTUs. The function of HS and RS soil fungi was different and can classified into eight trophic modes(). Among these trophic modes, Pathotroph-Saprotroph-Symbiotroph was the most abundant fungal guild, followed by the Pathotroph and Saprotroph. Very few symbiotrophs were observed. Although many OTUs were not assigned a trophic mode during FUNGuild analysis, there was still a significantly increase in Pathotrophs in the RS soil compared with HS soil samples. However, Saprotrophs showed the reverse trend, and were more abundant in the HS soil. The results indicated that the microbial functional profiles of the HS and RS soil was obviously different.
3.6. Correlation analysis of relative high abundance microbiomes and soil physicochemical properties
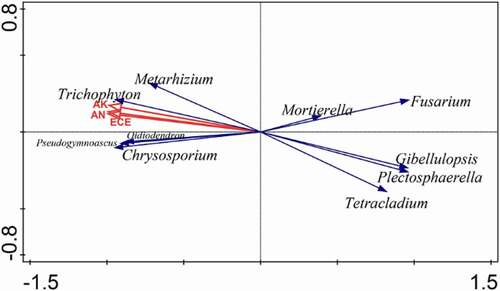
In order to explore the dominant factors affecting the microbiomes of American ginseng HS and RS soil, the RDA analysis of high relative abundance microbiomes and soil physicochemical properties was performed at the genus level, and reanalysis was performed on the basis of effects(). The adjusted interpretation of variance was 91.6% (Table S2). The results showed that soil CEC (p = .032, F = 31.9) had a significant effect on the American ginseng RS soil microbiomes, and the interpretation rate was 88.9%. Moreover, CEC was negatively correlated with Fusarium, Plectosphaerella and Gibellulopsis, whereas it was correlated positively with Oidiodendron and Metarhizium.
4. Discussion
4.1. Soil physiochemical characteristics of the rot root of American ginseng
Soil pH has significantly impact on crop growth and microbial community composition in the soil.Citation36,,Citation37 In our study, the pH of American ginseng RS soil significantly decreased compared with the HS group(), which was in line with a former study that the pH of Panax ginseng(the same genus of American ginseng) planting soil with dangerous rusty root phenomenon decreases significantly.Citation26 American ginseng in northern China usually grows in acid soil, but the lower pH value may affect the healthy growth of American ginseng. Additionally, the growth of American ginseng has high requirements for various substances in the soil. Phosphorus is an essential component of nuclear and membrane structure in plants. Normal phosphorus levels can complete the healthy metabolism of protein in plants, stimulate the growth of plant roots, and increase the absorption of mineral nutrients by rhizomes, to alleviate the damage of plant diseases.Citation38,Citation39 In our results, the RS soil phosphorus significantly decreased compared with HS soil. Furthermore, potassium can promote the development of the thick outer wall of epidermal cells, thus preventing the occurrence of diseases.Citation40,Citation41 As shown in , the RS soil had significantly less potassium than the HS rhizosphere soil. Further, plants obtain their nutrition from two natural sources: organic matter and minerals.Citation42 Soil organic matter consist of any plant or animal material that returns to the soil and undergoes decomposition. It provided nutrients and habitat for soil organisms, organic matter can also condense soil particles into aggregates and improve soil water retention capacity.Citation43 Soil organic carbon, a component of soil organic matter, can affect soil properties, which linked to crop yield.Citation44 The HS soil had a significantly high soil organic matter and soil organic carbon content compared to the RS soil. Overall, the decrease of some nutrients in RS soil may be associated with the rot root of American ginseng.
4.2. Soil microbiomes characteristics of American ginseng rot root
In this study, we compared the composition of fungal communities between soils of HS and RS American ginseng using Illumina MiSeq high-throughput sequencing. Overall, the α-diversity analysis of RS and HS soil, Chao 1, ACE and Sob index values for fungi were lower in RS soil of American ginseng than in those of HS soil. We then performed β-diversity analysis using unweighted UniFrac, Jaccard and Bray-Curtis distances. The unweighted UniFrac, Jaccard and the Bray-Curtis distance are the index to determine the difference of species composition in different soil samples. They can calculate the characteristics of the composition of different species in the sample. Jaccard only considers the presence or absence of species in the sample, while Bray-Curtis only considers the presence or absence of species and the relative abundance of different species in samples.Citation45 Our findings on fungal diversity are consistent with previous findings that the microbial diversity in healthy plant soils is greater than that in diseased plant soils.Citation46 There were significant differences between RS group and HS group, indicating that soil microorganisms in RS group had significant changes compared with HG group.
Further analysis uncovered a strong imbalance in the composition of fungal microbial communities between the soils of HS and RS of American ginseng, which was mainly due to differences in the dominant genera and their relative abundances. Moreover, LEfSe showed that six fungal genera were more abundant in the soil of RSt American ginseng than that of healthy one (p < .05, LDA >3.5). Soil microbial diversity and community composition play an essential role in maintaining the function, health and quality of soil ecosystem,Citation47 while the occurrence of medicinal plant diseases is mainly controlled by the imbalance of soil microbial diversity and community composition.Citation48 Because pathogens cause rot root in American ginseng and contribute to crop losses at the time of harvest, various studies have investigated the relationship between soil microbes and rot root;Citation19,Citation49,Citation50 however, most studies have considered only one or two pathogenic fungi that are involved in the occurrence of rot root in American ginseng, such as Ditylenchus destructor,Citation51 Phytophthora cactorumCitation52 and Cylindrocarpon destructans.Citation53
In our study, LEfSe indicated that six fungal genera, namely, Gibellulopsis, Fusarium, Plectosphaerella, Tetracladium, Gibberella and Ilyonectria, were considerably more abundant in the soil samples of RS of American ginseng than in those of HS soil (p < .05). Generally, three of these six genera (Ilyonectria, Plectosphaerella and Fusarium)Citation21,Citation54,Citation55have been widely reported to be closely associated with the occurrence of rot root in medicinal plants of American ginseng. However, interstingly, Gibellulopsis and Gibberella which can cause rot root of American ginseng have not been reported. As far as we known, Gibellulopsis chrysanthemi is responsible for seedling rot of garland chrysanthemum, and the symptoms (light brown spots first appeared on lower leaves of seedlings, and the leaves blighted or rotted.) are similar to those of ginseng rot root.Citation56 Gibberella can lead to the ear rot of corn, which called Gibberella ear rot.Citation57 Severe rot of leaves, peduncles and flowers caused by Gibberella zeae was also found on potted plants of hyacinth.Citation58 To date, there have been no reports of Tetracladium being associated with plant diseases. The soil microbial community contains many pathogenic, nonpathogenic, and symbiotic microorganisms that interact with plant roots simultaneously.Citation59 The relative abundance of rot root pathogens of American ginseng increased significantly, such as Ilyonectria, Plectosphaerella and Fusarium, may co-act on American ginseng. In view of these observations, this study shows for the first time that the increase in the relative abundance of Gibellulopsis and Gibberella in the soil of American ginseng may be related to the occurrence of rot root in this plant. More research is needed to test this hypothesis and identify the strains.
The HTS platform was employed to analyze the abundance and richness of low-abundance microbial species in environmental samples, overcoming the isolation limitations of culture-based methods. Most microorganisms cannot be cultured using traditional culture techniques.Citation60 Xia et al. identified 55 fungal genera from Chinese Cordyceps by Illumina Miseq sequencing, but these genera were not observed using culture-dependent approach.Citation61 In our study, we first used HTS to discover the potential fungi associated with rot root of American ginseng, providing an fast and rapid approach for the discovery of pivotal fungi associated with plant disease.
4.3. Correlation analysis of relative high abundance microbiomes and soil physicochemical properties
RDA analysis results shows that soil CEC was the dominant factor affecting the soil microbiomes. CEC refers to the total amount of various cations (e.g., K+, Na+, Ca2+, Mg2+, NH4+, H+ and Al3+) that can be adsorbed by negatively charged soil colloid. The value is expressed in terms of the amount of the various ions in the soil per kilogram, cmol/kg,Citation62 which is often used as an index to evaluate soil fertilizer-preserving ability.Citation63 ECE was negatively correlated with Fusarium, Plectosphaerella and Gibellulopsis, whereas it was correlated positively with Oidiodendron and Metarhizium. Ginseng roots growing in the field are susceptible to several soil-borne diseases. These diseases are primarily caused by several species of FusariumCitation21and Plectosphaerella.Citation55 However, several Oidiodendron species have long been recognized as typical ericoid mycorrhizal fungal partners.Citation64 Mycorrhizae fungi have an ecologically significant role in the establishment of the host plant in heathlands by facilitating the transfer of nutrition to the host plant and by contributing to the detoxification of the root environment.Citation65 Furthermore, Metarhizium not only kill pest insects also boosts plant growth by providing nitrogenous nutrients and increasing resistance to plant pathogens.Citation66,Citation67 In summary, CEC is the major factor that need to be considered for rot root of American ginseng, because it was positively correlated with beneficial microbiomes and negatively correlated with pathogens.
5. Conclusion
This study was the first to reveal two pivotal microbiomes associate with rot root of American ginseng by high-throughput sequencing. Moreover, the major factor affecting the soil microbiomes was explored. The following conclusions were obtained: (1) The soil physicochemical properties and fungal community structures of rot root American ginseng and healthy root significantly differed. (2) The relative abundances of several pathogenic fungi, such as Ilyonectria, Plectosphaerella and Fusarium, were significantly higher enriched in the soils of rot root of American ginseng than that in healthy ginseng.(3) This study was the first to highlight that a significant increase in the relative abundances of Gibellulopsis and Gibberella, may be associated with the rot root American ginseng. (4) Soil cation exchange capacity was identified as the important factor in shaping microbial communities between the healthy root and rot root. Our study is of great significance for the biocontrol of rot root, and management of medicinal plant cultivation.
Supplemental Material
Download MS Word (301.9 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Wang YP, Choi HK, Brinckmann JA, Jiang X, Huang LF. Chemical analysis of panax quinquefolius (North American ginseng): a review. J Chromatogr A. 2015;1426:1–10. doi:https://doi.org/10.1016/j.chroma.2015.11.012.
- Wu Q, Song JY, Sun YQ, Suo FM, Li CJ, Luo HM, Liu Y, Li Y, Zhang X, Yao H, et al. Transcript profiles of Panax quinquefolius from flower, leaf and root bring new insights into genes related to ginsenosides biosynthesis and transcriptional regulation. Physiol Plant. 2010;;138(2):134–149.https://doi.org/10.1111/j.1399-3054.2009.01309.x.
- Anderson RC, Fralish JS, Armstrong JE, Benjamin PK. The ecology and biology of panax-quinquefolium-l (Araliaceae) in illinois. Am Midl Nat. 1993;;129(2):357–372. doi:https://doi.org/10.2307/2426517.
- Ren G, Chen F. Simultaneous quantification of ginsenosides in American ginseng (Panax quinquefolium) root powder by visible/near-infrared reflectance spectroscopy. J Agric Food Chem. 1999;;47:2771–2775. https://pubs.acs.org/doi/10.1021/jf9812477.
- Jovanovski E, Dascalu A, Jenkins A, Sievenpiper JL, Vuksan V. American ginseng from five different sources has differential effects on postprandial blood glucose. Faseb J. 2006;;20:1019–A1019. https://faseb.onlinelibrary.wiley.com/doi/10.1096/fasebj.20.5.A1019-a
- Kim HY, Kang KS, Yamabe N, Nagai R, Yokozawa T. Protective effect of heat-processed American ginseng against diabetic renal damage in rats. J Agri Food Chem. 2007;;55(21):8491–8497. doi:https://doi.org/10.1021/jf071770y.
- Mucalo I, Rahelić D, Jovanovski E, Bozikov V, Romić Z, Vuksan V. Effect of American ginseng (Panax quinquefolius L.) on glycemic control in type 2 diabetes. Coll Antropol. 2012;;36:1435–1440. https://www.researchgate.net/publication/235420922
- Yoo KM, Lee C, Lo YM, Moon B. The hypoglycemic effects of American red ginseng (Panax quinquefolius L.) on a diabetic mouse model. J Food Sci. 2012;;77(7):147–152. doi:https://doi.org/10.1111/j.1750-3841.2012.02748.x.
- Durairaj P, Breda M, Miller SC. Quantitative augmentation of immune cells in elderly normal mice by short-term, daily consumption of an extract of North American ginseng (Panax quinquefolius). Biomed Res-India. 2013;;24:199–205. https://www.researchgate.net/journal/Biomedical-Research-0970-938X
- Guerrero-Analco JA, Azike CG, Romeh AA, Charpentier PA, Pei H, Lui EMK, Arnason JT. Bioactive Polysaccharides of North American Ginseng Panax quinquefolius L. in modulation of immune function: preliminary chemical and biological characterization. Planta Med. 2013;;79(10):835–836. doi:https://doi.org/10.1055/s-0033-1348593.
- Yu X, Yang X, Cui B, Wang L, Ren G. Antioxidant and immunoregulatory activity of alkali-extractable polysaccharides from North American ginseng. Int J Biol Macromol. 2014;;65:357–361. doi:https://doi.org/10.1016/j.ijbiomac.2014.01.046.
- Wang LJ, Yao Y, Sang W, Yang XS, Ren GX. Structural features and immunostimulating effects of three acidic polysaccharides isolated from panax quinquefolius. Int J Biol Macromol. 2015;;80:77–86. doi:https://doi.org/10.1016/j.ijbiomac.2015.06.007.
- Wang MQ, Guilbert LJ, Ling L, Li J, Wu YQ, Xu S, Pang P, Shan JJ. Immunomodulating activity of CVT-E002, a proprietary extract from North American ginseng (Panax quinquefolium). J Pharm Pharmacol. 2001;;53(11):1515–1523. doi:https://doi.org/10.1211/0022357011777882.
- Lian XY, Zhang Z, Stringer JL. Anticonvulsant and neuroprotective effects of ginsenosides in rats. Epilepsy Res. 2006;;70(2–3):244–256. doi:https://doi.org/10.1016/j.eplepsyres.2006.05.010.
- Wang JY, Yang JY, Wang F, Fu SY, Hou Y, Jiang B, Ma J, Song C, Wu CF. Neuroprotective effect of pseudoginsenoside-F11 on a rat model of parkinson’s disease induced by 6-hydroxydopamine. J evidence-based complementary. Altern Med. 2013;2013:152798. http://downloads.hindawi.com/journals/ecam/2013/152798
- Wang CZ, Zhang ZY, Wan JY, Zhang CF, Anderson S, He X, Yu C, He T-C, Qi L-W, Yuan C-S, et al. Protopanaxadiol, an active ginseng metabolite, significantly enhances the effects of fluorouracil on colon cancer. Nutrients. 2015;;7(2):799–814.https://doi.org/10.3390/nu7020799.
- Jung Y, Kim K, Bian Y, Ngo T, Bae ON, Lim KM, Chung JH. Ginsenoside Rg3 disrupts actin-cytoskeletal integrity leading to contractile dysfunction and apoptotic cell death in vascular smooth muscle. Food Chem Toxicol. 2018;;118:645–652. doi:https://doi.org/10.1016/j.fct.2018.06.015.
- Yang L, Hou A, Zhang J, Wang S, Man W, Yu H, Zheng S, Wang X, Liu S, Jiang H, et al. Panacis quinquefolii radix: a review of the botany, phytochemistry, quality control, pharmacology, toxicology and industrial applications research progress. Front Pharmacol. 2020;;11:602092. https://doi.org/10.3389/fphar.2020.602092
- DesRochers N, Walsh JP, Renaud JB, Seifert KA, Yeung KKC, Sumarah MW. Metabolomic profiling of fungal pathogens responsible for root rot in American ginseng. Metabolites. 2020;;10(1):35. doi:https://doi.org/10.3390/metabo10010035.
- Ji XL, Hou CY, Shi MM, Yan YZ, Liu YQ. An insight into the research concerning panax ginseng C. A. Meyer polysaccharides: a review. Food Rev Int. 2020;;5:1–17. https://www.tandfonline.com/doi/full/10.1080/87559129.2020.1771363
- Jiao X, Lu X, Chen AJ, Luo Y, Hao JJ, Gao W. Effects of fusarium solani and F. oxysporum infection on the metabolism of ginsenosides in American ginseng roots. Molecules. 2015;;20(6):10535–10552. doi:https://doi.org/10.3390/molecules200610535.
- Kong W, Wei R, Logrieco AF, Wei J, Wen J, Xiao X, Yang M. Occurrence of toxigenic fungi and determination of mycotoxins by HPLC-FLD in functional foods and spices in China markets. Food Chem. 2014;;146:320–326. doi:https://doi.org/10.1016/j.foodchem.2013.09.005.
- Al-Hindi RR, Aly SE, Hathout AS, Alharbi MG, Al-Masaudi S, Al-Jaouni SK, Harakeh SM. Isolation and molecular characterization of mycotoxigenic fungi in agarwood. Saudi J Bio Sci. 2018;;25(8):1781–1787. doi:https://doi.org/10.1016/j.sjbs.2017.07.008.
- Tedersoo L, Bahram M, Polme S, Koljalg U, Yorou NS, Wijesundera R, Ruiz LV, Vasco-Palacios AM, Thu PQ, Suija A, et al. Fungal biogeography. global diversity and geography of soil fungi. Science. 2014;;346(6213):1256688.https://doi.org/10.1126/science.1256688.
- Wei X, Wang X, Cao P, Gao Z, Chen AJ, Han J. Microbial community changes in the rhizosphere soil of healthy and rusty panax ginseng and discovery of pivotal fungal genera associated with rusty roots. BioMed Res Int. 2020;2020:8018525. doi:https://doi.org/10.1155/2020/8018525.
- Bian X, Xiao S, Zhao Y, Xu Y, Yang H, Zhang L. Comparative analysis of rhizosphere soil physiochemical characteristics and microbial communities between rusty and healthy ginseng root. Sci Rep. 2020;;10(1):15756. doi:https://doi.org/10.1038/s41598-020-71024-8.
- Wu Z, Hao Z, Sun Y, Guo L, Huang L, Zeng Y, Wang Y, Yang L, Chen B. Comparison on the structure and function of the rhizosphere microbial community between healthy and root-rot Panax notoginseng. Appl Soil Ecol. 2016;;107:99–107. doi:https://doi.org/10.1016/j.apsoil.2016.05.017.
- Huang LF, Song LX, Xia XJ, Mao WH, Shi K, Zhou YH, Yu JQ. Plant-soil feedbacks and soil sickness: from mechanisms to application in agriculture. J Chem Ecol. 2013;;39(2):232–242. doi:https://doi.org/10.1007/s10886-013-0244-9.
- Toju H, Tanabe AS, Yamamoto S, Sato H, Lespinet O. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS One. 2012;;7(7):e40863. doi:https://doi.org/10.1371/journal.pone.0040863.
- Ankenbrand MJ, Keller A, Wolf M, Schultz J, Forster F. ITS2 database V: twice as much. Mol. Biol. Evol. 2015;;32(11):3030–3032. doi:https://doi.org/10.1093/molbev/msv174.
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;;19(9):1639–1645. doi:https://doi.org/10.1101/gr.092759.109.
- Caporaso JG QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010; 7: 335–336.https://www.nature.com/articles/nmeth.f.303
- W C. HW ggplot2: an implementation of the grammar of graphics R package version 0.7. URL: htp:/CRAN.R-project. org/package=ggplot2. 2008.https://cran.r-project.org/web/packages/ggplot2/index.html
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;;12(6):60. doi:https://doi.org/10.1186/gb-2011-12-6-r60.
- NH N, Song Z, ST B, Branco S, Tedersoo L, Menke J, JS S, Kennedy PG. FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016;;20:241–248. doi:https://doi.org/10.1016/j.funeco.2015.06.006.
- Wan W, Tan J, Wang Y, Qin Y, He H, Wu H, Zuo W, He D. Responses of the rhizosphere bacterial community in acidic crop soil to pH: changes in diversity, composition, interaction, and function. Sci Total Environ. 2020;;700:134418. doi:https://doi.org/10.1016/j.scitotenv.2019.134418.
- Guo JH, Liu XJ, Zhang Y, Shen JL, Han WX, Zhang WF, Christie P, Goulding KWT, Vitousek PM, Zhang FS, et al. Significant acidification in major Chinese croplands. Science. 2010;;327(5968):1008–1010.https://doi.org/10.1126/science.1182570.
- Warren SLJH. Mineral nutrition of crops: fundamental mechanisms and implications. Hort Science. 2004;;39:462.
- Dordas C. Role of nutrients in controlling plant diseases in sustainable agriculture:a review. Agron Sustainable Dev. 2008;;28(1):33–46. doi:https://doi.org/10.1051/agro:2007051.
- Zhang H, Zeng Z, Zou Z, Zeng F. Climate, life form and family jointly control variation of leaf traits. Plants (Basel). 2019;;8:8080286. https://www.ncbi.nlm.nih.gov/pubmed/31416214.
- Sharma S, Duveiller E, Basnet R, Karki CB, Sharma RC. Effect of potash fertilization on helminthosporium leaf blight severity in wheat, and associated increases in grain yield and kernel weight. Field Crop Res. 2005;;93:142–150. htpp:www.elsevier.com/locate/fcr/10.1016/j.fcr.2004.09.016.
- Khosro M. Soil management, microorganisms and organic matter interactions: a review. Afr J Bio Technol. 2011;;10. https://www.mendeley.com/catalogue/6f06e277-a6d1-39b9-bc29-ce8dfd2cf619/
- Johnston AE, Poulton PR, Coleman K. Soil organic matter: its importance in sustainable agriculture and carbon dioxide fluxes. Adv Agron. 2009:1–57. https://www.sciencedirect.com/science/article/pii/S0065211308008018
- Martinez E, Fuentes J, Acevedo E. Soil organic carbon and soil properties. revista de la ciencia del suelo y nutricion. Vegetal. 2008;;8:68–96. http://dx.doi.org/10.4067/S0718-27912008000100006.
- Meyer F, Bremges A, Belmann P, Janssen S, McHardy AC, Koslicki D. Assessing taxonomic metagenome profilers with OPAL. Genome Biol. 2019;;20(1):51. doi:https://doi.org/10.1186/s13059-019-1646-y.
- Xu L, Ravnskov S, Larsen J, Nilsson RH, Nicolaisen M. Soil fungal community structure along a soil health gradient in pea fields examined using deep amplicon sequencing. Soil Biol Biochem. 2012;;46:26–32. doi:https://doi.org/10.1016/j.soilbio.2011.11.010.
- Garbeva P, van Veen JA, van Elsas JD. Microbial diversity in soil: selection microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol. 2004;;42(1):243–270. doi:https://doi.org/10.1146/annurev.phyto.42.012604.135455.
- Wu L, Wang J, Huang W, Wu H, Chen J, Yang Y, Zhang Z, Lin W. Plant-microbe rhizosphere interactions mediated by Rehmannia glutinosa root exudates under consecutive monoculture. Sci Rep. 2015;;5(1):15871. doi:https://doi.org/10.1038/srep15871.
- Rahman M, Punja ZK. Factors influencing development of root rot on ginseng caused by Cylindrocarpon destructans. Phytopathology. 2005;;95:1381–1390. https://doi.org/10.1094/PHYTO-95-1381.
- Cabral A, Groenewald JZ, Rego C, Oliveira H, Crous PW. Cylindrocarpon root rot: multi-gene analysis reveals novel species within the Ilyonectria radicicola species complex. Mycol Prog. 2011;;11(3):655–688. doi:https://doi.org/10.1007/s11557-011-0777-7.
- ZG Z, ZH W. First report of root rot of american ginseng (panax quinquefolium) caused by itylenchus destructor in China. Plant Dis. 2007;;91(4):459. doi:https://doi.org/10.1094/PDIS-91-4-0459C.
- Li TSC, Utkhede RS, Wardle DA. Chemical and biological control of leaf blight and root rot caused by Phytophthora cactorum in American ginseng. Can J Plant Pathol. 1997;;19(3):297–300. doi:https://doi.org/10.1080/07060669709500527.
- Farh ME, Kim YJ, Kim YJ, Yang DC. Cylindrocarpon destructans/Ilyonectria radicicola-species complex: causative agent of ginseng root-rot disease and rusty symptoms. J Ginseng Res. 2018;;42(1):9–15. doi:https://doi.org/10.1016/j.jgr.2017.01.004.
- Jiang Y, Ran C, Chen L, Yin W, Liu Y, Chen C, Gao J. Purification and characterization of a novel antifungal flagellin protein from endophyte bacillus methylotrophicus NJ13 against ilyonectria robusta. Microorganisms. 2019;;7(7120605):605. doi:https://doi.org/10.3390/microorganisms7120605.
- Han L, Zhou X, Zhao Y, Wu L, Ping X, He Y, Peng S, He X, Du Y. First report of Plectosphaerella plurivora causing root rot disease in Panax notoginseng in China. J Phytopathol. 2020;;168:375–379. https://onlinelibrary.wiley.com/doi/abs/10.1111/jph.12901.
- Kawaradani M, Taguchi K, Okada K, Hirooka Y, Sato T. Seedling rot of garland chrysanthemum caused by gibellulopsis chrysanthemi and ecological characters of the causal fungus. J Gen Plant Pathol. 2013;;79(5):346–349. doi:https://doi.org/10.1007/s10327-013-0462-6.
- Kawakami A, Kato N, Sasaya T, Tomioka K, Inoue H, Miyasaka A, Hirayae K. Gibberella ear rot of corn caused by fusarium asiaticum in Japan. J Gen Plant Pathol. 2015;;81(4):324–327. doi:https://doi.org/10.1007/s10327-015-0593-z.
- Tomioka K, Hirooka Y, Aoki T, Sato T. Fusarium rot of hyacinth caused by Gibberella zeae (anamorph: fusarium graminearum). J Gen Plant Pathol. 2008;;74(3):264–266. doi:https://doi.org/10.1007/s10327-008-0088-2.
- De Coninck B, Timmermans P, Vos C, Cammue BPA, Kazan K. What lies beneath: belowground defense strategies in plants. Trends Plant Sci. 2015;;20(2):91–101. doi:https://doi.org/10.1016/j.tplants.2014.09.007.
- Guo M, Jiang W, Luo J, Yang M, Pang X. Analysis of the fungal community in ziziphi spinosae semen through high-throughput sequencing. Toxins (Basel). 2018;;10(10):494. doi:https://doi.org/10.3390/toxins10120494.
- Xia F, Chen X, Guo MY, Bai XH, Liu Y, Shen GR, Li YL, Lin J, Zhou XW. High-throughput sequencing-based analysis of endogenetic fungal communities inhabiting the Chinese Cordyceps reveals unexpectedly high fungal diversity. Sci Rep. 2016;;6(1):33437. doi:https://doi.org/10.1038/srep33437.
- Boeva NM, Bocharnikova YI, Belousov PE, Zhigarev VV. Determining the cation exchange capacity of montmorillonite by simultaneous thermal analysis method. Russ J Phys Chem A. 2016;;90(8):1525–1529. doi:https://doi.org/10.1134/S0036024416080057.
- Jiang L, Geng ZC, Li SS, She D, He XS, Zhang Q, Liang C,Liu XD,Jing WM,Wang SL. Soil cation exchange capacity and exchangeable base cation content in the profiles of four typical soils in the xi-shui forest zone of the qilian mountains. Acta Ecologica SinicaSoil. 2012;;32(11):3368–3377.https://doi.org/10.5846/stxb201104280563.
- Zhang C, Yin L, Dai S. Diversity of root-associated fungal endophytes in rhododendron fortunei in subtropical forests of China. Mycorrhiza. 2009;;19(6):417–423. doi:https://doi.org/10.1007/s00572-009-0246-1.
- Baba T, Hirose D, Sasaki N, Watanabe N, Kobayashi N, Kurashige Y, Karimi F, Ban T. Mycorrhizal formation and diversity of endophytic fungi in hair roots of vaccinium oldhamii miq. in Japan. microbes environ. 186-9. 2016;;31. https://www.ncbi.nlm.nih.gov/pubmed/27297892.
- KMS A, Hue SM. Mode of infection of metarhizium spp. fungus and their potential as biological control agents. J Fungi. 2017;3:jof3020030. https://www.ncbi.nlm.nih.gov/pubmed/29371548.
- Lira AC, Mascarin GM, Delalibera Junior I. Microsclerotia production of metarhizium spp. for dual role as plant biostimulant and control of spodoptera frugiperda through corn seed coating. Fungal Biol. 2020;124(8):689–699. doi:https://doi.org/10.1016/j.funbio.2020.03.011.