ABSTRACT
Autophagy is an evolutionarily conserved pathway for the degradation of damaged or toxic components. Under normal conditions, autophagy maintains cellular homeostasis. It can be triggered by senescence and various stresses. In the process of autophagy, autophagy-related (ATG) proteins not only function as central signal regulators but also participate in the development of complex survival mechanisms when plants suffer from adverse environments. Therefore, ATGs play significant roles in metabolism, development and stress tolerance. In the past decade, both the molecular mechanisms of autophagy and a large number of components involved in the assembly of autophagic vesicles have been identified. In recent studies, an increasing number of components, mechanisms, and receptors have appeared in the autophagy pathway. In this paper, we mainly review the recent progress of research on the molecular mechanisms of plant autophagy, as well as its function under biotic stress and abiotic stress.
1. Introduction
In the process of growth and development, plants inevitably encounter various adverse conditions, such as drought, high salinity, pathogen invasion, and nutrient deficiency. Therefore, plants need a sophisticated protein degradation mechanism to cope with various stresses and maintain cell homeostasis.Citation1 The ubiquitin (Ub)/26S proteasome system (UPS) is a primary proteolytic pathway for abnormal and short-lived proteins in plants.Citation2 However, some large particles, such as protein complexes, protein aggregates and damaged organelles, need to be degraded by autophagy. Autophagy can be divided into three categories: chaperone-mediated autophagy, microautophagy, and macroautophagy.Citation3,Citation4 Chaperone-mediated autophagy is a selective autophagy process that has been reported only in mammals.Citation5 This process is independent of vesicles; it recruits individual target proteins through specific chaperone proteins and promotes their direct entry into lysosomes for degradation.Citation6 In the process of microautophagy, cytoplasmic substances converge to the surface of the vacuole and are engulfed by invagination of the vacuolar membrane. Then, the tonoplast pinches off to release autophagic bodies, which are intravacuolar vesicles containing cytoplasmic components. Macroautophagy is one of the most well-known autophagic processes and is hereinafter referred to as autophagy. Once autophagy is activated, the phagophore forms and expands, eventually closing to form double membrane vesicles called autophagosomes.Citation7,Citation8 Then, mature autophagosomes are delivered to lysosomes (mammals) or vacuoles (yeast and plants) and degraded by acidic hydrolases in these lytic compartments.Citation9,Citation10
In the 1990s, a group of genes needed for autophagosome formation were identified in yeast (Saccharomyces cerevisiae) through forward genetics; these are called core ATG genes.Citation11–14 Most of the core ATG proteins are conserved in yeast, mammals and plants and can be divided into four groups: (i) the ATG1/ATG13 kinase complex; (ii) the phosphatidylinositol 3-kinase (PI3K) complex; (iii) the ATG9 cycling system; and (iv) the ATG8-PE (phosphatidylethanolamine) and ATG12-ATG5 conjugation systems.Citation15–17 Although ATG genes are similar in yeast and plants, some plant ATG homologs are found in the form of multigene families. In Arabidopsis (Arabidopsis thaliana), the ATG1 family contains four genes, the ATG4, ATG12 and ATG13 families each contain two genes, and the ATG18 and ATG8 families contain eight and nine genes, respectively.Citation18–20 Different ATG8 proteins may have different functions in plants, similar to those observed in mammals.Citation21 The molecular mechanisms of autophagy are sophisticated and involve many ATG proteins; among them, the activity of the ubiquitin-like protein ATG8 is crucial for autophagy. Since ATG8 is attached to the membrane of the phagophore, autophagosome and autophagic body, it is often used as a standard marker in eukaryotic cells.Citation21,Citation22
In plants, transcriptional and epigenetic regulation of autophagy has been well reviewed.Citation23,Citation24 The transcriptional regulation of ATG genes is extremely important for autophagy to maintain cellular homeostasis under adverse environmental conditions. The transcription factors (TFs) including WRKY33, HY5, HsfA1a, ERF5, BZR1 are involved in this process.Citation25–29 Based on the yeast one-hybrid assay, it has been identified that 225 TFs from 35 families are able to bind to the promoters of 4 ATG8 genes. Meanwhile, these TFs participate in plant development and environmental stress response.Citation30 These results lay a solid foundation for us to understand the transcriptional regulation of plant autophagy. In addition, epigenetic changes such as DNA methylation, histone modification and noncoding RNAs also affect the expression of ATG genes as well as the process of autophagy.Citation24
In this review, we mainly summarize the current understanding of autophagic mechanisms in plants and supplement the latest research progress. On the other hand, the effects of autophagy on plant nutrition metabolism, growth and development and its strong relationship with various biotic and abiotic stresses are discussed.
2. Molecular Mechanisms of Autophagy in Plants
Autophagy is a conserved degradation process in plants, mammals and yeast. There are many brilliant reviews that have summarized the functions of related components and molecular mechanisms of autophagy in plants in detail.Citation5,Citation6,Citation31,Citation32 In this section, we systematically review the plant autophagy mechanism and add some recent research results ().
Figure 1. Schematic diagram of known or proposed steps in macroautophagy. Upon starvation, SnRK1 inhibits TOR activity, which causes ATG13 dephosphorylation and promotes the combination of ATG1, ATG13, ATG101 and ATG11 to initiate autophagy. ATG9, together with ATG18 and ATG2, facilitate the delivery of lipids to the extending phagophore. PI3K complexes synthesize PI3P and decorate the phagophore with it. Subsequently, ATG8 is modified with PE and then conjugated to the phagophore. Once formed, autophagosomes are transported to the vacuole, potentially using the microtubule cytoskeleton controlled by the ESCRT machinery. Finally, autophagosomes fuse with the tonoplast through the v-SNARE-type mechanism and release autophagic bodies into the vacuolar lumen for degradation.
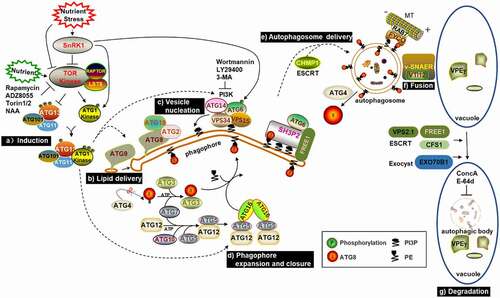
2.1. Induction of Autophagy
Autophagy occurs when environmental and developmental signals affect the activity of several crucial protein kinases. In plants, the heterotrimeric complex sucrose nonfermenting 1-related protein kinase1 (SnRK1) is a major metabolic regulator in response to energy and nutrition deficiency.Citation33,Citation34 The energy sensor SnRK1 consists of one catalytic α-type subunit (KIN10 and KIN11 in Arabidopsis) and two noncatalytic β-type and γ-type subunits.Citation35,Citation36 In Arabidopsis, the kin10 kin11 double mutant could not survive, and overexpression of KIN10 not only delayed leaf senescence and floweringCitation37 but also increased autophagosome formation and tolerance to nutritional deficiency,Citation38 indicating that KIN10 regulates development and autophagy in plants. Sucrose nonfermenting 1 (Snf1) and AMP-activated kinase (AMPK) are orthologs of SnRK1 in yeast and animals, respectively. They are important energy and metabolic sensors that maintain homeostasis and activate the autophagy pathway during nutrient deficiency.Citation39,Citation40 More importantly, they can regulate autophagy through target of rapamycin (TOR).Citation41,Citation42 TOR is a significant negative regulator of autophagy, and this kinase has two effectors, namely, lethal with sec thirteen 8 (LST8) and RAPTOR. When nutrition is sufficient, TOR is in the active state, which prevents the assembly of the ATG1/ATG13 kinase complex by phosphorylating ATG13. When nutrition is deficient, TOR is in the inactive state, which causes ATG13 dephosphorylation and promotes the combination of ATG1, ATG13 and other factors, such as ATG101 and ATG11.Citation20,Citation43,Citation44 TOR deletion mutants result in embryo death,Citation45 while downregulation of TOR expression results in inhibition of plant growth and autophagy.Citation46,Citation47 When plants are exposed to various stresses, autophagy can be activated by TOR-independent or TOR-dependent pathways, and auxin acts upstream of TOR.Citation48
Through the study of various mutants of the ATG1 kinase complex, researchers confirmed that there are two autophagic signaling pathways in Arabidopsis under fixed-carbon stress. Although ATG1 kinase is essential for autophagic activation during short-term fixed-carbon starvation as well as nitrogen deprivation, it is not necessary for autophagic activation induced by long-term fixed-carbon starvation. The second pathway seems to maintain strong autophagy through the direct phosphorylation of ATG6 by KIN10 in the downregulation or deficiency of the ATG1 kinase complex.Citation49 Little is known about the functions of Rho GTPase signaling in plant autophagy. Recently, it was found that Sec5, a subunit of the exocyst complex, could bind to activated ROP8 under stress conditions, and the ROP8-Sec5 module could further transduce upstream ROP8 signaling and promote autophagosome formation by recruiting the ATG1 and PI3K complex to the phagophore. However, at which step the Sec5 exocyst complex functions in plant autophagy is still unknown. One possibility is that Sec5 activates autophagy by recruiting ATG1/13 complexes. Another possibility is that Sec5 plays a targeting role during autophagosome progression.Citation50
2.2. Lipid Delivery, Vesicle Nucleation, Phagophore Expansion and Closure
After the completion of ATG1/ATG13 kinase complex assembly, the subsequent steps of autophagy are promoted. ATG9, together with ATG18 and ATG2, facilitate the delivery of essential lipids to the extending phagophore.Citation51,Citation52 Unlike other ATG proteins, ATG9 is a special multispanning membrane protein and plays an important role in the formation of autophagosomes.Citation53 Recently, the 3D structure of ATG9 at 7.8 Å resolution was presented in Arabidopsis. This result has provided us with structural insights into the oligomeric mode of ATG9, identified the important interaction interfaces, and preliminarily assigned the transmembrane helices located in the membrane-embedded region.Citation54 Compared with atg5 and atg7 mutants, autophagy is not seriously blocked in Arabidopsis atg9 mutants.Citation19 Moreover, upon autophagic induction in the atg9 mutant, an autophagy-related tubular structure accumulates and associates with the membrane of the endoplasmic reticulum (ER).Citation52 PI3K complexes synthesize phosphatidylinositol 3-phosphate (PI3P) and decorate the phagophore with it. The PI3K complex is composed of complex I and complex II, both of which contain three core proteins: vacuolar protein sorting 34 (VPS34), VPS15 and ATG6/VPS30/Beclin1. The difference is that complex I contains ATG14, whereas complex II includes VPS38.Citation55 VPS38 is not necessary for autophagy in Arabidopsis.Citation56 Arabidopsis mutants lacking three core proteins of PI3K are sterile;Citation57,Citation58 moreover, the atg14a atg14b mutants have typical autophagy-deficient phenotypes, and vps38 mutants show a severe phenotype. Compared with atg14a atg14b and vps38, the phenotype of atg14a atg14b vps38 is severely compromised but still accumulates a low level of PI3P. However, this mutant is sensitive to wortmannin, which indicates that the three core subunits of the PI3K complex are adequate to synthesize the right amount of PI3P.Citation55 In Arabidopsis, after treatment with wortmannin and analysis of atg double mutants, ATG11, ATG9 and PI3K were found to act upstream of ATG2. Plant ATG1 and PI3K complexes function in the initiation of autophagy, while ATG2 plays a role in the later stage of autophagic vesicle biogenesis.Citation59 Subsequently, starvation-induced ATG8 is modified with PE and then conjugated to the autophagosome precursor (phagophore).Citation60 In the Arabidopsis genome, there are two homologs of the ATG4 gene, termed ATG4a and ATG4b.Citation19 ATG4 is a cysteine (Cys) protease that can cleave the C-terminus of an inactive precursor of ATG8 to expose the conserved glycine (Gly) residue.Citation61 ATP-dependent ATG7 is an E1-activating enzyme that activates ATG8 and binds the Gly residue of ATG8 with its Cys residue by a thioester linkage. Then, ATG7 donates ATG8 to the Cys residue of the E2-conjugating enzyme ATG3. With the help of the E3 ligase complex, ATG8 attaches to PE and localizes to the phagophore.Citation62 The other Ub-fold protein ATG12 is also activated by ATG7 and transferred to its specific E2-conjugating enzyme ATG10. Then, ATG12, through its C-terminal Gly, covalently attaches to the specific Lys of ATG5. The dimeric protein ATG16 combines with the ATG12-ATG5 conjugate to form a hexameric complex that is associated with the phagophore and facilitates ATG8 lipidation.Citation63–65
3D imaging analysis of growing phagophores in living Arabidopsis tissue showed that ATG5 plays a decisive role in the formation of autophagosomes. After autophagy is induced, ATG5-GFP is localized on the outer surface of the ER, and ATG8 is recruited to the initial phagophore. Then, the phagophore expands into a nearly two-dimensional cisterna, whose edge is associated with ATG5. When it expands into a cup-shaped structure, ATG5 becomes a ring-like structure to encircle the aperture of the phagophore. Finally, once the phagophore is sealed, ATG5 will leave the structure, and the autophagosome will also detach from the ER membrane.Citation66 SH3 domain-containing protein 2 (SH3P2) is a typical non-ATG protein in Arabidopsis. By tracing the SH3P2-GFP fusion protein, it was found that SH3P2 is mainly located in phagophore assembly site (PAS) derived from omegasome-like structures or isolated cup-shaped double-membrane structures.Citation67,Citation68 When autophagy is activated, SH3P2 transfers to PAS and promotes phagophore curvature by binding to PIP, PI3K complex and ATG8. More importantly, RNAi knockdown of SH3P2 inhibits autophagosome formation.Citation67 After the autophagosome matures, the ATG8 proteins lining the outer membrane are released by ATG4 protease for recycling, whereas ATG8 proteins lining the inner membrane enter the vacuole and are degraded along with the cargo.Citation61,Citation69
2.3. Is the Cytoskeleton Involved in the Formation of Autophagosomes?
The cytoskeletal components seem to be essential for the formation of autophagosomes in plants. It has been shown that Arabidopsis ATG8 could bind to microtubules in vitro.Citation70 Tobacco (Nicotiana benthamiana) protein Joka2, which is a member of a family of selective autophagy cargo receptors, can colocalize with microfilaments as well as microtubules.Citation71 ATG6 can colocalize and interact with microtubules in tobacco, and the destruction of microtubules reduces the formation of autophagosomes.Citation72 The SCAR/WAVE complex is essential for the actin-related protein 2/3 (ARP2/3) complex to control the dynamics and assembly of filamentous actin and promote actin nucleation.Citation73,Citation74 All of the members of the SCAR/WAVE complex also exist in Arabidopsis.Citation75,Citation76 Nck-associated protein 1 (NAP1) is a membrane-localized protein of the SCAR/WAVE complex,Citation77 and NAP1-GFP puncta can be triggered by constant pression. These NAP1 puncta not only associate with the cytoskeleton and ER but also colocalize with ATG8. More importantly, the nap1 mutant is sensitive to nutritional deficiency.Citation78 However, it is unknown whether the involvement of NAP1 in autophagy relies on its actin nucleation activity. Studies have shown that the effect of the cytoskeleton on autophagy varies across model systems. In plants, actin filaments are unexpectedly dispensable for bulk autophagy. However, actin filaments may be involved in other unknown types of autophagy or selective autophagy.Citation79 Arabidopsis selective autophagy cargo receptor neighbor of breast cancer 1 (NBR1) is a homolog of mammalian p62 and NBR1. It was found that YFP-AtNBR1 not only exhibited cytosolic and punctate patterns but also showed microtubule patterns especially in the cortical layer. Instead of microfilament depolymerizer Latrunculin B, treatments with microtubule depolymerizer Oryzalin disrupted the microtubule localization and influenced the vacuolar delivery of YFP-AtNBR1 upon autophagy induction. This suggests that AtNBR1 may need the assistance of microtubules to shuttle its goods to vacuoles during plant autophagy.Citation80 Therefore, it seems that microtubules play a more important role than microfilaments in the autophagic pathway, and the exact relationship between autophagy and cytoskeleton remains to be further studied.
2.4. The Delivery of Autophagosomes
The transport of autophagosomes to vacuoles may involve several special trafficking mechanisms. The endosomal sorting complex required for transport (ESCRT) machinery is an evolutionarily conserved protein complex that is mainly responsible for sorting ubiquitinated protein entry into the intraluminal vesicles (ILVs) of prevacuolar compartments/multivesicular bodies (PVCs/MVBs), which then fuse with lytic organelles to degrade cargos.Citation81,Citation82 As a component of ESCRT, FYVE domain protein required for endosomal sorting 1 (FREE1) participates in vacuolar protein transport and the regulation of autophagic degradationCitation83,Citation84 In addition, FREE1 directly interacts with SH3P2 to regulate the fusion progress between autophagosomes and vacuoles. Both autophagosomes and ATG8-PE accumulate to a certain extent in the free1 mutant.Citation83 It has been reported that ESCRT-III subunit VPS2.1 and cell death-related endosomal FYVE/SYLF protein 1 (CFS1) are also related to transport and fusion.Citation85,Citation86 The deubiquitinating enzyme AMSH3 can interact with ESCRT-III, and loss of AMSH3 affects vacuolar transport and vacuole formation.Citation87 AMSH1 is involved in autophagy mediated degradation. More importantly, it interacts with VPS2.1, and both mutants have autophagy-related phenotypes.Citation85 On the other hand, the plant exocyst component EXO70B1 is also necessary for autophagosome formation and transport. The exo70B1 mutants are susceptible to nitrogen deficiency. Furthermore, the amounts of intravacuolar autophagic bodies are decreased in this mutant.Citation88
2.5. Fusion of Autophagosomes and Tonoplasts
Finally, autophagosomes fuse with the tonoplast through the vesicular soluble N-ethylmaleimide-sensitive factor attachment protein receptor (v-SNARE)-type mechanismCitation89 and release autophagic bodies into the vacuolar lumen for degradation. As a member of the VTI family of v-SNAREs, VTI12 may be involved in plant autophagy. The vti12 mutant grows normally under nutrient-rich conditions, but senescence is accelerated under nutrient-poor conditions.Citation89 AtVPS45 and VTI12 may be functionally related because both mutants exhibit trafficking defects, and autophagy-related defects are found in VPS45-silenced lines.Citation90 In animals, some small Rab GTPases bind to autophagosome membranes to regulate the fusion process of lysosomes and autophagosomes by recruiting distinctive effector proteins. As one of the members, Rab7 plays an important role in the maturation of autophagosomes and endosomes and their subsequent fusion step with lysosomes (animals) or vacuoles (yeast).Citation91–93 RabG3b, a homologous protein of Rab7 in Arabidopsis, is located in ATG8e-labeled autophagic structures. During tracheary element (TE) differentiation, autophagy is activated, and transgenic plants overexpressing a constitutively active RabG3b (RabG3bCA) accumulate many autophagic structures. RabG3b may regulate TE differentiation as well as TE programmed cell death (PCD) through autophagy.Citation94 The hypersensitive response (HR) is a core component of the plant defense response to pathogens. RabG3b can facilitate HR cell death through autophagy. In Arabidopsis RabG3bCA cells, autophagosomes also increase during HR cell death.Citation95 In Arabidopsis, whether Rab7 can regulate the fusion process between autophagosomes and vacuoles remains to be further studied, and the function of Rab GTPase in autophagosome biogenesis is still unclear. Recently, it has been reported that a specific Rab GTPase, AtRabD2a, can specifically interact with ATG1 complex and the coat protein complex II (COPII) machinery and act as a link between autophagy and COPII.Citation96,Citation97
2.6. Degradation of Autophagic Bodies in Vacuoles
Autophagic bodies can be degraded into amino acids and other small molecules for recycling by various hydrolases in the vacuole.Citation98 Most enzymes in vacuoles are synthesized as precursors that need to be processed into active forms. Vacuolar processing enzyme-γ (VPEγ) is a cysteine proteinase that can be induced during plant senescence and death. VPEγ degrades cargos by releasing pro-proteins into the acidic vacuole lumen to initiate processing.Citation99 The continuous acidification of vacuoles is extremely important for the activity of hydrolases. Vacuolar-type ATPases (V-ATPases) are ATP-dependent proton pumps that are located at the tonoplast and continuously pump H+ into the vacuole to maintain its acidic environment.Citation100,Citation101 Concanamycin A (ConcA), a crucial inhibitor of V-ATPase, can prevent the acidification of vacuoles and inhibit the degradation of autophagic bodies, thus increasing its visibility under the microscope.Citation61,Citation102 The use of this inhibitor provides a very important approach for scientific research on autophagy. Recently, it has been reported that the cytoplasmic soluble GFP was accumulated in vacuoles after treatment with ConcA.Citation103 Further studies showed that GFP signal colocalized with the autophagic marker mCherry-ATG8f in vacuoles, but free GFP did not display direct interaction with ATG8s. Therefore, when reporting specific cytosol components degraded by selective autophagy, colocalization analysis need to be carried out in the cytoplasm rather than in the vacuole, so as to avoid the nonselective transport of cytoplasmic components into the vacuole. In addition, other parallel experiments need to be combined to judge whether cytoplasmic proteins are degraded by selective autophagy, which will contribute to identify the cargos and receptors of selective autophagy in plants.Citation103
3. The Function of Autophagy in Plants
After years of research, we have a profound understanding of the functions of autophagy in plants ( and ). Under normal conditions, autophagy occurs at basal levelsCitation104,Citation105 and participates in a variety of biotic and abiotic stresses.Citation31,Citation106,Citation107 In Arabidopsis, most atg mutants grow normally under nonstress conditions and show a greater sensitivity to nutritional deficiency. The rosette leaves and cotyledons of the atg mutants show early senescence under short-day (SD) conditions, and the seed setting rate is lower than that of the wild type (WT).Citation21 In addition, ATG genes are upregulated at the transcriptional level under carbon and nitrogen deficiency.Citation108,Citation109,Citation110
Table 1. Summary of physiological functions of autophagy related components and some regulators in plants
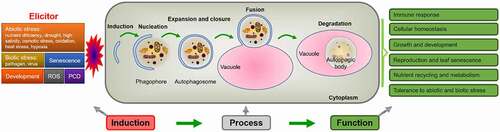
Figure 2. Simplified scheme of the induction, process and function of autophagy in plants. Autophagy is induced by various factors, through a series of processes such as nucleation, expansion, closure, fusion and degradation, so as to recycle various nutrients and realize diverse physiological functions.
3.1. Nutrient Recycling and Metabolism
In recent years, many studies have shown that autophagy plays a key role in nutrient cycling and reuse. As the primary carbon source of plant respiration, carbohydrates are mainly stored in chloroplasts in the form of starch for nighttime consumption.Citation111 Autophagy is active at night, and the content of starch in the leaves of autophagy-deficient Arabidopsis and tobacco is increased.Citation112 In Arabidopsis, the growth inhibition of atg mutants under SD will be relieved by the application of exogenous sucrose or treatment with full light conditions, and the starchless and atg double mutants have severe starvation phenotypes under SD, indicating that autophagy is involved in energy availability.Citation113 Lipids and proteins will be used as backup carbon sources when carbohydrates are deficient in cells.Citation114 For example, autophagy regulates nutritional supplements and lipid metabolism in anthers, and it is critical for anther development after meiosis in the tapetum cells of rice (Oryza sativa).Citation115 In Arabidopsis, it has been found that basal autophagy facilitates the synthesis of triacylglycerol, while inducible autophagy promotes the degradation of lipid droplets. These results indicate that autophagy plays a dual role in regulating plant lipid metabolism.Citation116 Autophagy has an effect on lipid homeostasis under dark-induced senescence.Citation117 The statistical characterization of metabolic data indicates that autophagy deficiency has noteworthy effects on metabolite profiles in Arabidopsis leaves. Compared with the WT, 4 metabolites in Arabidopsis atg5 mutants were largely decreased, including urate, saccharopine, putrescine and N6,N6,N6-trimethyllysine, while 36 metabolites were increased. These results suggest that autophagic proteolysis is related to plant metabolic processes.Citation118 Various omics analyses have shown that autophagy has a crucial effect on metabolism and leads to a morphological phenotype during starvation in Arabidopsis. Delayed growth is found in etiolated atg mutants, and the content of free amino acids also decreases.Citation119 Autophagy is not only involved in amino acid cycling but also responsible for increasing the accumulation of proteogenic dipeptides during recovery from heat stress in Arabidopsis.Citation120 In maize (Zea mays), multi-omics analysis clearly identifies the role of autophagy in various cellular processes under nitrogen-replete and -starvation conditions, and shows that autophagic recycling has a significant impact on lipid turnover and proteome remodeling.Citation121 Maize multi-omics also showed that autophagy plays remarkable roles in nucleotide, amino acid, and carbohydrate metabolism during fixed-carbon starvation.Citation122
In higher plants, nitrogen is the most important nutrient element and plays a decisive role in plant productivity.Citation123 Autophagy also affects nitrogen remobilization, and it has been confirmed that nitrogen use efficiency (NUE) decreases at the whole-plant level and is independent of senescence in autophagy mutants.Citation124 Moreover, rosette leaves show the accumulation of several proteins due to aberrant protein degradation in the Arabidopsis atg mutant.Citation125 Although leaf senescence of Osatg7-1 is significantly advanced, the nitrogen concentration in leaves is still high, indicating that the NUE and biomass production of Osatg7-1 are lower than those in WT.Citation126 In soybean (Glycine max) callus cells, the constitutive expression of GmATG8c could enhance tolerance to nitrogen deficiency, and GmATG8c overexpressing Arabidopsis could survive under long-term nutrient deficiency. In a suitable growing environment, the overexpression lines grew faster than the WT and had larger inflorescences and higher seed yields.Citation127 Studies have shown that overexpression of ATG8 has a beneficial effect on nitrogen remobilization at the whole-plant level without affecting plant biomass or total seed yield in Arabidopsis.Citation128 Consistently, compared with the WT, most Arabidopsis autophagy mutants are hypersensitive to nitrogen starvation,Citation20,Citation51,Citation55,Citation102,Citation129,Citation130 which further proves that autophagy regulates nitrogen cycling. In contrast to the decrease in free amino acids in etiolated seedlings,Citation119 it was found that the content of ammonium and free amino acids in the atg mutant increased significantly at 60 days after sowing (DAS).Citation125,Citation131 The output of ammonium and amino acids requires strong phloem loading, so the longevity of vein tissue is necessary.Citation132 It is possible that the mutant leaves at 60 DAS affect phloem loading during senescence, resulting in the accumulation of ammonium and total amino acids.Citation125 The metabolome data from atg mutants showed that amino acid interconversion metabolism mainly regulates oxidative stress through various pathways and ultimately affects the concentration of amino acids.Citation131
3.2. Growth and Development
To ensure survival, many kinds of developing seeds synthesize a large number of storage proteins, which are degraded during early germination to start photosynthesis and synthesize new plant organs.Citation133,Citation134 During seed development, synthetic storage proteins are transported from the ER to protein storage vacuoles through a process similar to autophagy.Citation21,Citation135,Citation136 The expression levels of many ATG genes are upregulated during seed maturation as well as drying and approach the highest level in dry seeds in Arabidopsis.Citation21,Citation137 In Arabidopsis, ATG8f is specifically expressed in seeds and siliques. Seed development is accelerated and storage protein accumulation is altered in atg5 mutants.Citation138 The ATG8-PE adducts are increased in starchy endosperm during seeds maturation after pollination in maize, and many ATG8-PE adducts are detected after seed germination, indicating that autophagy is beneficial to the remobilization of nutrients in endosperm during early seed development.Citation139 RNA-seq analysis shows that the transcription abundance of the maize ATG gene increased in the endosperm, indicating that autophagy is involved in its maturation and death process during seed development.Citation140 Autophagy is very important for grain filling in maize and Arabidopsis.Citation124,Citation140 ATI1 and ATI2 are special ATG8-interacting proteins in Arabidopsis. They are partially associated with the ER membrane network under suitable conditions, and with spherical compartments under carbon starvation. In the presence of the exogenous hormone abscisic acid (ABA), the expression levels of ATI1 and ATI2 will affect the germination rate in Arabidopsis seeds.Citation141
AtATG18a RNAi plants are not only sensitive to nutrient deficiency but also show premature leaf senescence. Monodansylcadaverine (MDC) staining showed that AtATG18a RNAi plants could hardly produce autophagosomes in response to nutritional starvation.Citation142 Autophagy is beneficial to the process of proteolysis of stromal proteins, such as Rubisco activase, Rubisco large subunit and chloroplast glutamine synthetase. In Arabidopsis atg5 and atg7 mutants, the degradation of these proteins is incomplete.Citation143 The degradation of starch in leaves also requires the assistance of autophagy during the night. Similar to ATG6-silenced tobacco, the starch degradation ability of other ATG-silenced plants shows a decrease in varying degrees.Citation112 When E-64d was used to treat Arabidopsis root tip cells, almost no vacuolar inclusions were observed in the atg2 and atg5 mutants, but a large number of vacuolar inclusions appeared in the atg9 mutant. These results indicate that not only ATG2 but also ATG5 are essential for autophagy in Arabidopsis roots, while ATG9 is helpful but not essential. An autophagy inhibitor called 3-methyladenine could inhibit the accumulation of this vacuolar inclusion.Citation105 As an important component of autophagy, the small GTPase RabG3b may regulate TE differentiation via activating autophagy.Citation94,Citation144 In Arabidopsis, autophagy could facilitate glucose-mediated root meristem maintenance by modulating reactive oxygen species (ROS) levels.Citation145 Arabidopsis PTEN, a special lipid and protein dual phosphatase homologous to animal PTENs (phosphatase and tensin homologs deleted on chromosome 10), regulates autophagy in pollen tubes by interfering with the dynamics of PI3P.Citation146 During Arabidopsis pollen development, PI3K plays a pivotal role in vacuole reorganization and nuclear division.Citation58 The components of PI3K complex, including AtVPS34, AtVPS15, AtATG6, participated in pollen germination.Citation57,Citation58,Citation147–150 OsATG7 knockout mutants not only had no accumulation of starch grains and lipid bodies in pollen grains but also showed male sterility, limited anther dehiscence and decreased pollen germination activity.Citation115 Autophagy is necessary for the regulation of gene expression and timely progression of tapetal PCD during rice pollen maturation.Citation151 Autophagy also plays an important role in pollen development and tapetum degeneration during heat-induced tapetal PCD abortion.Citation152 In tobacco, pollen germination is accompanied by the improvement of autophagy activity and compartmental cytoplasmic deletion is vital for male fertility.Citation153 The role of autophagy in plant male reproductive development has been well summarized in some reviews.Citation154,Citation155
3.3. Immune Response
Unlike mammals, plants lack mobile immune cells, so they can only depend on the innate immune system to recognize pathogenic nonself molecules for defense. Plants resist the invasion of pathogenic microorganisms and maintain their normal growth and development through two immune defense systems: pathogen-associated molecular pattern-triggered immunity (PTI) and effector-triggered immunity (ETI). In general, ETI can lead to cell death in host plants, referred to as HR,Citation156,Citation157 a form of PCD.
The most prominent phenotype of autophagy mutants is premature senescence accompanied by accelerated cell death.Citation107 It has been reported that autophagy possesses both pro-survival and pro-death roles and may be connected with the plant defense response, especially ETI.Citation158 HR cell death may play a significant role in plant disease resistance by restricting the spread of pathogens. The most significant characteristic of HR is the accelerated death of cells in the local area around the infection. For example, tobacco protein NbATG6, an ortholog of the yeast and mammalian ATG6/VPS30/ BECLIN 1, could limit HR PCD to the infection site. In the HR PCD reaction, both infected cells and uninfected cells could induce autophagy, which is weakened in ATG6-deficient plants, indicating that autophagy plays an important role in innate immunity and negatively regulates PCD.Citation159 In AtATG6 antisense (AtATG6-AS) plants, HR PCD induced by avirulent Pst DC3000 carrying the AvrRpm1 effector protein could spread from infected sites to uninfected areas. In addition, the disease-related cell death caused by highly virulent Pst DC3000 bacterial infection is also partially misregulated in AtATG6-AS plants.Citation160 In plant innate immunity, there are several examples to prove that autophagic components contribute greatly to cell death. By detecting HR PCD in the Arabidopsis atg mutants, it was found that only a special subset of immune receptors participate in the autophagy pathway for HR execution.Citation161,Citation162
Autophagy contributes to plant immunity against diverse pathogens, and the mechanisms are quite different. Even in response to the same pathogen, there are different proteins involved in autophagy by different mechanisms. In Arabidopsis, the resistance of atg5, atg10 and atg18a mutants to necrotrophic fungal pathogens is impaired, but the resistance to biotrophic bacterial invaders is enhanced. Therefore, autophagy, as a pro-death or anti-death function, largely depends on the infection strategy as well as the lifestyle of invasive microorganisms.Citation163 ATG7-dependent autophagy forms an anti-death plant mechanism to regulate the containment of cell death and immunity to necrotrophic fungal infection. It has been suggested that ATG7 plays an important role in plant resistance to fungal pathogens.Citation164 WRKY33 is an essential transcription factor for plant resistance to necrotrophic pathogens in Arabidopsis. ATG18a and WRKY33 could interact with each other, and both mutants are highly sensitive to the necrotrophic pathogen Botrytis cinerea, which not only induces autophagy gene expression but also promotes autophagosome formation. WRKY33- and jasmonate-mediated signaling pathways and autophagy may synergistically regulate the plant defense responses to necrotrophic pathogens.Citation25
When Arabidopsis responds to avirulent bacterial pathogens, RabG3b facilitates HR cell death by activating autophagy, and this process is partially ATG5 dependent.Citation95 BAG6 belongs to the Bcl-2-associated athanogene (BAG) family, whose cleavage leads to plant defense and autophagy.Citation165 In Arabidopsis, glyceraldehyde-3-phosphate dehydrogenase (GAPCs) could regulate cell death when inoculated with Pseudomonas syringae.Citation166 Similarly, ATG3 interacts with GAPCs to regulate PCD and autophagy during innate immunity in tobacco.Citation167 BAK1 is an important regulator of plant resistance that can interact with and phosphorylate ATG18a. Phosphorylation of ATG18a can inhibit the formation of autophagosomes and the subsequent transfer to vacuoles, resulting in a decrease in autophagy activity and resistance against Botrytis cinerea.Citation168 In Arabidopsis, exocyst subunit Exo70B2 interacts with ATG8 and kinase MPK3. In addition, phosphorylation of Exo70B2 by MPK3 enhances the interaction between MPK3 and ATG8. Phosphonull variants exhibit higher ETI and hypersensitivity to benzothiadiazole (BTH), which result in autophagy and secretion.Citation169
Salicylic acid (SA) is an important plant defense hormone that can effectively resist biotrophic pathogens in living plant tissues.Citation170–172 Studies have shown that pathogen-induced HR PCD and early senescence in atg mutants are related to SA signaling,Citation173,Citation174 and SA largely accumulates in atg mutants.Citation163,Citation173 In addition, pathogen infection and BTH treatment induce ATG gene expression,Citation25,Citation160,Citation173 all of which indicate that there is a negative feedback loop between SA signaling and autophagy to control each other. Furthermore, some studies have shown that autophagy can function SA-dependently and -independently.Citation173,Citation174 Plant age has a significant impact on the spread of SA-dependent HR cell death in autophagy mutants. For a specific host-pathogen interaction, autophagy only has a pro-death role in a limited area during a particular period (possibly transiently and early), while autophagy plays a pro-survival role in most other periods (especially in senescent cells and tissues).Citation161,Citation173,Citation175,Citation176
Autophagy is the key part of plant immunity and the crucial target of pathogens. Plant pathogens regulate host autophagy by injecting effector proteins. However, the exact molecular mechanism of autophagy manipulated by plant pathogens remains unclear. Recent studies have systematically analyzed the interaction between ATG protein of Arabidopsis and effectors of various plant pathogens and verified the interactions of a subset of effectors with biochemical and in vivo experiments. The results show that effectors of plant pathogens regulate plant autophagy through a variety of mechanisms.Citation177,Citation178 PexRD54, an effector from the Irish potato famine pathogen Phytophthora infestans, binds host ATG8CL to trigger the formation of autophagosomes. PexRD54 could deplete the autophagic cargo receptor Joka2 from ATG8CL complexes and hamper the positive role of Joka2 in pathogen defense.Citation179 Additionally, the structural basis of this interaction has been analyzed.Citation180
Increasing evidence shows that autophagy also plays a significant role in the process of plant virus infection, and an excellent review has been published.Citation181 During viral infection, autophagy is thought to exert both antiviral and proviral roles.Citation181,Citation182 Autophagy plays an antiviral role when plants are infected by DNA virusesCitation159,Citation183–187 or RNA viruses.Citation188–190 Discoveries of the antiviral effect of selective autophagy have attracted much attention.Citation186,Citation189,Citation190 In turn, plant viruses can hijack, subvert, or even exploit the autophagy pathway for transmission or infection.Citation188,Citation191 However, we still know little about the role of autophagy in the plant antiviral defense response, and there is no report indicating that plant viruses are completely limited by autophagy. How autophagy is inhibited in particular plant-virus interactions needs to be further studied.
3.4. Abiotic Stress
In Arabidopsis, abiotic stresses such as nutrient deficiency, drought, heat stress, oxidation and salt can induce autophagy () and most atg mutants are highly sensitive to these adverse conditions.Citation106,Citation142,Citation192–194 Autophagy is an important mechanism for plant stress response. It has been reported that many ATG genes are involved in the abiotic stress response ().
Drought and high salinity are unavoidable abiotic stresses for plants. Drought can not only cause membrane damage but also decrease the activity of cytoplasmic and organelle proteins and even lead to complete denaturation. In wild emmer wheat (Triticum dicoccoides), the expression pattern of ATG8 shows that ATG8 is extremely induced under drought stress.Citation195 Drought treatment can enhance the activity of five genes, ATG4a, 4b, 8a, 8g and 8h, and prolong the effect on ATG4a and 4b in wheat (Triticum aestivum L.).Citation196 In tomato (Solanum lycopersicum), HsfA1a regulates drought tolerance by activating autophagy, and autophagy facilitates plant survival by degrading insoluble proteins produced under drought stress. HsfA1a can positively regulate drought-induced autophagy and directly bind to the ATG10 and ATG18f promoters to regulate their expression.Citation27 In Arabidopsis, the sensitivity of KIN10-OE lines to drought and hypoxia is lower than that of the WT.Citation38
High salinity can cause ion stress and osmotic stress and may lead to plant death. The main ions involved in salt stress are H+, Na+, K+ and Ca2+, and intracellular homeostasis is maintained by the interaction of these ions.Citation197 In Arabidopsis, both wortmannin-treated plants and pi3K mutants are extremely sensitive to salt.Citation198 The expression of the GFP-AtATG8f-HA fusion protein seems to make plants more sensitive to mild osmotic stress as well as salt stress.Citation199 OsATG10b mutants have a greater sensitivity to both methyl viologen (MV) and high salt treatment.Citation200 Compared with the wild type, autophagy-defective RNAi-AtATG18a plants are hypersensitive to drought and salt.Citation106 Under high salt conditions, TaATG8a has the highest transcript accumulation among the three TaATG8s.Citation196 In Arabidopsis, atg mutants are extremely sensitive to osmotic and salt stress, and ATG8-OX plants show better germination than WT plants on NaCl-containing plates.Citation201
Heat stress is a common and important factor limiting plant productivity. It can lead to the accumulation of denatured proteins, which easily aggregate.Citation202 In tomato, heat-induced autophagy may be cooperatively regulated by ATG and WRKY33 proteins through selective degradation of protein aggregates. In addition, ATG5- and ATG7-silenced plants showed increased sensitivity to heat stress.Citation203 Recent studies have shown that autophagy plays a specific role in resetting the memory of heat stress in plants.Citation204 In Arabidopsis, ATG2 could be induced by high temperature. Moreover, the mRNA levels of ATG6, ATG5, ATG18a and ATG12a in WT plants are significantly increased after high temperature treatment.Citation205 Transcriptome analysis of Arabidopsis WT and atg5 mutant plants prior to and after heat stress showed that autophagy has little effect on heat shock gene expression induced by heat, but autophagy plays an active role in the expression of numerous other stress- and defense-related genes in the early stage of plant heat stress response. These findings provide significant new insights into the extensive functions of autophagy.Citation206 When the amount of unfolded or misfolded proteins within the ER increases beyond the capacity of the intracellular folding and degradation system under heat stress and other adverse conditions, it will eventually lead to ER stress.Citation207
NADPH oxidase inhibitors could impede autophagy induction under salt stress and nutrient starvation, but not under osmotic stress, indicating that there are NADPH oxidase-dependent or -independent pathways for activating autophagy.Citation106 Compared with the WT, RNAi-AtATG18a transgenic plants were susceptible to MV treatment and accumulated more oxidative proteins on account of a lower degradation rate. When ConcA was applied, oxidized proteins were observed in WT root cells but not RNAi-AtATG18a root cells. These results indicate that the oxidative proteins induced by oxidative stress can be degraded by autophagy.Citation192 In addition, overexpression of ATG5 and ATG7 can enhance the resistance of plants to oxidative stress.Citation208 In mammals, both hypoxia and ROS can cause autophagy.Citation209 In Arabidopsis, submergence and ethanol treatments can induce the accumulation of ROS in the atg mutants more than in the WT, the ROS signaling pathways and SA-induced defense may be involved in the regulation of autophagy-related hypoxia response.Citation210 Various stresses may induce ROS production, which in turn activates autophagy.Citation176 In contrast, ROS significantly accumulated in the leaves of atg2 and atg5 mutants grown in SD for 8 weeks, indicating that autophagy can partially limit ROS levels.Citation173 It has been reported that autophagy can control ROS homeostasis by removing oxidized peroxisomes, thus optimizing stomatal opening.Citation211 In summary, these results indicate that autophagy components play an important role in plant tolerance to abiotic stress.
Autophagy was originally defined as a nonselective bulk degradation process. However, in recent years, accumulated evidence has shown that the recruitment of targets into autophagosomes is highly selective.Citation5,Citation212 ATG8 interacts with diverse adaptor/receptor proteins to recruit specific targets for degradation by selective autophagy. These interactors usually contain an ATG8-interacting motif (AIM, or LC3-interacting region (LIR)) that contacts a hydrophobic patch on ATG8 collectively termed the LIR/AIM-docking site (LDS).Citation213 In Arabidopsis, RPN10 (regulatory particle non-ATPase 10) acts as an autophagy receptor for the ubiquitinated 26S proteasome; however, its binding to ATG8 rely on the ubiquitin-interacting motif (UIM) instead of the canonical AIM.Citation214 The UIM-docking site (UDS) is important for the interaction between RPN10 and ATG8.Citation215 UDS is located opposite to the LDS; thus, ATG8 can bind to AIM- and UIM-containing proteins simultaneously. The discovery of UIM-UDS interface greatly enlarges the range of autophagy adaptors and receptors, thereby expanding the reach of selective autophagy. There are several autophagy receptors in plants, some of which play a role in abiotic stress response ().
In Medicago truncatula, dehydrin MtCAS31 can interact with ATG8a. Under drought stress, MtCAS31 not only promotes autophagic degradation of aquaporin MtPIP2;7 but also reduces root hydraulic conductivity, thereby decreasing water loss and enhancing drought resistance.Citation216 COST1 is a plant-specific gene that promotes plant growth by inhibiting autophagy under suitable conditions. COST1 is degraded under drought conditions to activate autophagy and inhibit growth, thus enhancing drought tolerance.Citation217 DSK2 is a phosphorylation-regulated selective autophagy receptor. Due to tandem replication, there are two DSK2 paralogs genes (DSK2A and DSK2B) in Arabidopsis and both proteins are involved in BES1 degradation under drought stress conditions.Citation218 Arabidopsis TSPO, a tryptophan-rich sensory protein (TSPO)-related membrane protein,Citation219 its expression is induced by osmotic and salt stress.Citation219,Citation220 The degradation of TSPO is inhibited in atg5 mutant and is sensitive to PI3K inhibitor.Citation221 Arabidopsis ATG8-interacting proteins 1/2 (ATI1/2) are AIM-motif-containing proteins identified from yeast two-hybrid screen with ATG8f as bait.Citation141 ATI1 is involved in salt tolerance of Arabidopsis possibly by scavenging damaged plastid proteins.Citation222 Dicot-specific ATI3 proteins have a WxxL motif at the C-terminus, which is required for interaction between ATI3 and ATG8. Disruption of ATI3 significantly compromises plant heat tolerance.Citation223
Perhaps NBR1 is the most well-studied autophagy receptor in plants. Under heat stress, nbr1 mutants accumulate insoluble proteins. Ubiquitinated protein aggregates derived from denatured proteins are mainly degraded by NBR1-mediated autophagy. Like autophagy-deficient atg5 and atg7, nbr1 mutants exhibit hypersensitivity to heat, salt, oxidative and drought stresses.Citation193 Similarly, virus-induced gene silencing (VIGS) of NBR1 compromised tomato heat tolerance.Citation203 NBR1 also plays a key role as a receptor for selective autophagy during recovery from heat stress. It interacts with ROF1 (rotamase FKBP 1) and HSP90.1 (heat shock protein 90.1) and mediates their degradation by autophagy to regulate heat stress memory in Arabidopsis.Citation224 In tomato, studies have shown that BRs regulate NBR1-dependent selective autophagy in response to cold stress.Citation225 In poplar, PagNBR1 is also a significant selective autophagy receptor and confers salt tolerance by accelerating the activity of antioxidant system and autophagy.Citation226 In addition to the important role in these abiotic stresses, plant NBR1 is also involved in the regulation of plant-virus interactions and special protein secretion. And it may play a role in stress-induced pexophagy, sulfur nutrient responses and ABA signaling.Citation227,Citation228 Other selective autophagy receptors with very important functions have also been well summarized.Citation228 ATI3s, Sec62, Reticulons (Rtn) and C53 proteins are involved in ER-phagy. Adi3 and ORM1/2 are associated with immune responses. DSK2 and GSNOR1 are closely linked to signal transduction.Citation228,Citation229 RPN10 and PUXs (plant UBX domain-containing proteins) participate in proteaphagy and ERAD (ER-associated protein degradation), respectively.Citation228 The analysis of these crucial selective autophagy receptors and related autophagy pathways have provided significant new insights into plant responses under different stress conditions.
4. Conclusions and Prospects
Autophagy can effectively degrade damaged organelles and harmful proteins in cells, and it is very important for the recycling of nutrients. Although we already have a good understanding of the mechanism of plant autophagy and the core autophagy route is highly conserved in plants, there are still some important unknown factors inherent in the sophisticated molecular mechanisms and route of autophagy, and most studies of plant autophagy are based on mammals and yeast. Therefore, we need to further study whether traditional autophagy genes have new functions and whether there are new autophagy genes and mechanisms involved. Studies have shown that the autophagy system is very complex. On the one hand, the activity of autophagy is inevitably controlled by various stresses, such as nutrient deficiency, drought, salt, heat stress, oxidation, pathogens and viruses. On the other hand, the same ATG gene can also respond to diverse abiotic stresses. We should uncover how plants integrate various environmental signals to regulate autophagy. In addition, autophagy plays an important role in plant carbon and nitrogen metabolism as well as growth and development. More importantly, the study of autophagy in crops may increase the application of autophagy in agriculture, which can improve the resistance and yield of crops. Great progress in plant autophagy research in recent decades can be used to facilitate this process.
Abbreviations
Acknowledgments
This work was supported by the National Natural Science Foundation of China under Grant (31400220, 31970195); and the Fundamental Research Funds for the Central Universities under Grant (lzujbky-2021-kb05).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Vierstra RD. Proteolysis in plants: mechanisms and functions. Plant Mol Biol. 1996;32(1–2):1–17. doi:https://doi.org/10.1007/BF00039386.
- Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55(1):555–590. doi:https://doi.org/10.1146/annurev.arplant.55.031903.141801.
- Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118(1):7–18. doi:https://doi.org/10.1242/jcs.01620.
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21(22):2861–2873. doi:https://doi.org/10.1101/gad.1599207.
- Marshall, Richard S, Vierstra,Richard D. Autophagy: the master of bulk and selective recycling. Annu Rev Plant Biol. 2018;69:173–208. doi:https://doi.org/10.1146/annurev-arplant-042817-040606.
- Li F, Vierstra RD. Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012;17(9):526–537. doi:https://doi.org/10.1016/j.tplants.2012.05.006.
- Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14(12):759–774. doi:https://doi.org/10.1038/nrm3696.
- Yin Z, Pascual C, Klionsky DJ. Autophagy: machinery and regulation. Microb Cell. 2016;3(12):588–596. doi:https://doi.org/10.15698/mic2016.12.546.
- Takeshige K, Baba M, Tsuboi S, Ohsumi NY. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119(2):301–311. doi:https://doi.org/10.1083/jcb.119.2.301.
- Baba M, Takeshige K, Ohsumi BY. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol. 1994;124(6):903–913. doi:https://doi.org/10.1083/jcb.124.6.903.
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333(1–2):169–174. doi:https://doi.org/10.1016/0014-5793(93)80398-e.
- Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349(2):275–280. doi:https://doi.org/10.1016/0014-5793(94)00672-5.
- Harding TM. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131(3):591–602. doi:https://doi.org/10.1083/jcb.131.3.591.
- Klionsky DJ, Cregg JM, Dunn WA, Emr SD, Ohsumi Y. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5(4):539–545. doi:https://doi.org/10.1016/s1534-5807(03)00296-x.
- Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24(1):24–41. doi:https://doi.org/10.1038/cr.2013.168.
- Hurley JH, Young LN. Mechanisms of autophagy initiation. Annu Rev Biochem. 2017;86:225–244. doi:https://doi.org/10.1146/annurev-biochem-061516-044820.
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi:https://doi.org/10.1146/annurev-cellbio-092910-154005.
- Bassham DC, Laporte M, Marty F, Moriyasu Y, Ohsumi Y, Olsen LJ, Yoshimoto K. Autophagy in development and stress responses of plants. Autophagy. 2006;2(1):2–11. doi:https://doi.org/10.4161/auto.2092.
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129(3):1181–1193. doi:https://doi.org/10.1104/pp.011024.
- Suttangkakul A, Li F, Chung T, Vierstra RD. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell. 2011;23(10):3761–3779. doi:https://doi.org/10.1105/tpc.111.090993.
- Avin-Wittenberg T, Honig A, Galili G. Variations on a theme: plant autophagy in comparison to yeast and mammals. Protoplasma. 2012;249(2):285–299. doi:https://doi.org/10.1007/s00709-011-0296-z.
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12(9):823–830. doi:https://doi.org/10.1038/ncb0910-823.
- Yang C, Luo M, Zhuang X, Li F, Gao C. Transcriptional and epigenetic regulation of autophagy in plants. Trends Genet. 2020;36(9):676–688. doi:https://doi.org/10.1016/j.tig.2020.06.013.
- Cao JJ, Liu CX, Shao SJ, Zhou J. Molecular mechanisms of autophagy regulation in plants and their applications in agriculture. Front Plant Sci. 2021;11:618944. doi:https://doi.org/10.3389/fpls.2020.618944.
- Lai Z, Wang F, Zheng Z, Fan B, Chen Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011;66(6):953–968. doi:https://doi.org/10.1111/j.1365-313X.2011.04553.x.
- Yang C, Shen W, Yang L, Sun Y, Li X, Lai M, Wei J, Wang C, Xu Y, Li F, et al. HY5-HDA9 module transcriptionally regulates plant autophagy in response to light-to-dark conversion and nitrogen starvation. Mol Plant. 2020;13(3):515–531. doi:https://doi.org/10.1016/j.molp.2020.02.011.
- Wang Y, Cai S, Yin L, Shi K, Xia X, Zhou Y, Yu J, Zhou J. Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy. 2015;11(11):2033–2047. doi:https://doi.org/10.1080/15548627.2015.1098798.
- Zhu T, Zou L, Li Y, Yao X, Xu F, Deng X, Zhang D, Lin H. Mitochondrial alternative oxidase-dependent autophagy involved in ethylene-mediated drought tolerance in Solanum lycopersicum. Plant Biotechnol J. 2018;16(12):2063–2076. doi:https://doi.org/10.1111/pbi.12939.
- Wang Y, Cao JJ, Wang KX, Xia XJ, Shi K, Zhou YH, Yu JQ, Zhou J. BZR1 mediates brassinosteroid-induced autophagy and nitrogen starvation in tomato. Plant Physiol. 2019;179(2):671–685. doi:https://doi.org/10.1104/pp.18.01028.
- Wang P, Nolan TM, Yin Y, Bassham DC. Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy. 2020;16(1):123–139. doi:https://doi.org/10.1080/15548627.2019.1598753.
- Wang P, Mugume Y, Bassham DC. New advances in autophagy in plants: regulation, selectivity and function. Semin Cell Dev Biol. 2018;80:113–122. doi:https://doi.org/10.1016/j.semcdb.2017.07.018.
- Soto-Burgos J, Zhuang X, Jiang L, Bassham DC. Dynamics of autophagosome formation. Plant Physiol. 2018;176(1):219–229. doi:https://doi.org/10.1104/pp.17.01236.
- Sugden C, Crawford RM, Halford NG. Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5ʹ-AMP. Plant J. 1999;19(4):433–439. doi:https://doi.org/10.1046/j.1365-313x.1999.00532.x.
- Crozet P, Margalha L, Confraria A, Rodrigues A, Martinho C, Adamo M, Elias CA, Baena-González E. Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Front Plant Sci. 2014;5:190. doi:https://doi.org/10.3389/fpls.2014.00190.
- Polge C, Thomas M. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 2007;12(1):20–28. doi:https://doi.org/10.1016/j.tplants.2006.11.005.
- Emanuelle S, Hossain MI, Moller IE, Pedersen HL, van de Meene AM, Doblin MS, Koay A, Oakhill JS, Scott JW, Willats WG, et al. SnRK1 from Arabidopsis thaliana is an atypical AMPK. Plant J. 2015;82(2):183–192. doi:https://doi.org/10.1111/tpj.12813.
- Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448(7156):938–942. doi:https://doi.org/10.1038/nature06069.
- Chen L, Su ZZ, Huang L, Xia FN, Qi H, Xie LJ, Xiao S, Chen QF. The AMP-activated protein kinase KIN10 is involved in the regulation of autophagy in Arabidopsis. Front Plant Sci. 2017;8:1201. doi:https://doi.org/10.3389/fpls.2017.01201.
- Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25(18):1895–1908. doi:https://doi.org/10.1101/gad.17420111.
- Carroll B, Dunlop EA. The lysosome: a crucial hub for AMPK and mTORC1 signalling. Biochem J. 2017;474(9):1453–1466. doi:https://doi.org/10.1042/BCJ20160780.
- Lee JW, Park S, Takahashi Y, Wang HG. The association of AMPK with ULK1 regulates autophagy. PloS One. 2010;5(11):e15394. doi:https://doi.org/10.1371/journal.pone.0015394.
- Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, Choi AM, Chu CT, Codogno P, Colombo MI, et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36(13):1811–1836. doi:https://doi.org/10.15252/embj.201796697.
- Dobrenel T, Caldana C, Hanson J, Robaglia C, Vincentz M, Veit B, Meyer C. TOR signaling and nutrient sensing. Annu Rev Plant Biol. 2016;67:261–285. doi:https://doi.org/10.1146/annurev-arplant-043014-114648.
- Li F, Chung T, Vierstra RD. AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell. 2014;26(2):788–807. doi:https://doi.org/10.1105/tpc.113.120014.
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci U S A. 2002;99(9):6422–6427. doi:https://doi.org/10.1073/pnas.092141899.
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolai M, Bedu M, Robaglia C, Meyer C. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007;8(9):864–870. doi:https://doi.org/10.1038/sj.embor.7401043.
- Liu Y, Bassham DCTOR. Is a negative regulator of autophagy in Arabidopsis thaliana. PloS One. 2010;5(7):e11883. doi:https://doi.org/10.1371/journal.pone.0011883.
- Pu Y, Luo X, Bassham DC. TOR-dependent and -independent pathways regulate autophagy in Arabidopsis thaliana. Front Plant Sci. 2017;8:1204. doi:https://doi.org/10.3389/fpls.2017.01204.
- Huang X, Zheng C, Liu F, Yang C, Zheng P, Lu X, Tian J, Chung T, Otegui MS, Xiao S, et al. Genetic analyses of the Arabidopsis ATG1 kinase complex reveal both kinase-dependent and independent autophagic routes during fixed-carbon starvation. Plant Cell. 2019;31(12):2973–2995. doi:https://doi.org/10.1105/tpc.19.00066.
- Lin Y, Zeng Y, Zhu Y, Shen J, Ye H, Jiang L. Plant Rho GTPase signaling promotes autophagy. Mol Plant. 2021;14(6):905–920. doi:https://doi.org/10.1016/j.molp.2021.03.021.
- Xiong Y, Contento AL, Bassham DC. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005;42(4):535–546. doi:https://doi.org/10.1111/j.1365-313X.2005.02397.x.
- Zhuang X, Chung KP, Yong C, Lin W, Jiang L. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc Natl Acad Sci U S A. 2017;114(3):E426–E435. doi:https://doi.org/10.1073/pnas.1616299114.
- Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198(2):219–233. doi:https://doi.org/10.1083/jcb.201202061.
- Lai LTF, Yu C, Wong JSK, Lo HS, Benlekbir S, Jiang L, Lau WCY. Subnanometer resolution cryo-EM structure of Arabidopsis thalianaATG9. Autophagy. 2020;16(3):575–583. doi:https://doi.org/10.1080/15548627.2019.1639300.
- LiuF, Hu W, Li F, Marshall RS, Zarza X, Munnik T, Vierstra RD. AUTOPHAGY-RELATED14 and its associated phosphatidylinositol 3-kinase complex promote autophagy inArabidopsis. Plant Cell. 2020;32(12):3939–3960. doi:https://doi.org/10.1105/tpc.20.00285.
- Lee HN, Zarza X, Kim JH, Yoon MJ, Kim SH, Lee JH, Paris N, Munnik T, Otegui MS, Chung T. Vacuolar trafficking protein VPS38 is dispensable for autophagy. Plant Physiol. 2018;176(2):1559–1572. doi:https://doi.org/10.1104/pp.17.01297.
- Xu N, Gao XQ, Zhao XY, Zhu DZ, Zhou LZ, Zhang XS. Arabidopsis AtVPS15 is essential for pollen development and germination through modulating phosphatidylinositol 3-phosphate formation. Plant Mol Biol. 2011;77(3):251–260. doi:https://doi.org/10.1007/s11103-011-9806-9.
- Lee Y, Kim ES, Choi Y, Hwang I, Staiger CJ, Chung YY, Lee Y. The Arabidopsis phosphatidylinositol 3-kinase is important for pollen development. Plant Physiol. 2008;147(4):1886–1897. doi:https://doi.org/10.1104/pp.108.121590.
- Kang S, Shin KD, Kim JH, Chung T. Autophagy-related (ATG) 11, ATG9 and the phosphatidylinositol 3-kinase control ATG2-mediated formation of autophagosomes in Arabidopsis. Plant Cell Rep. 2018;37(4):653–664. doi:https://doi.org/10.1007/s00299-018-2258-9.
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2(3):211–216. doi:https://doi.org/10.1038/35056522.
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16(11):2967–2983. doi:https://doi.org/10.1105/tpc.104.025395.
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408(6811):488–492. doi:https://doi.org/10.1038/35044114.
- Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p–Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18(14):3888–3896. doi:https://doi.org/10.1093/emboj/18.14.3888.
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20(21):5971–5981. doi:https://doi.org/10.1093/emboj/20.21.5971.
- Kuma A. Formation of the 350-kDa Apg12-Apg5·Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem. 2002;277(21):18619–18625. doi:https://doi.org/10.1074/jbc.M111889200.
- Le Bars R, Marion J, Le Borgne R, Satiat-Jeunemaitre B, Bianchi MW. ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat Commun. 2014;5(10):4121. doi:https://doi.org/10.1038/ncomms5121.
- Zhuang X, Wang H, Lam SK, Gao C, Wang X, Cai Y, Jiang L. A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell. 2013;25(11):4596–4615. doi:https://doi.org/10.1105/tpc.113.118307.
- Zhuang X, Chung KP, Luo M, Jiang L. Autophagosome biogenesis and the endoplasmic reticulum: a plant perspective. Trends Plant Sci. 2018;23(8):677–692. doi:https://doi.org/10.1016/j.tplants.2018.05.002.
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151(2):263–276. doi:https://doi.org/10.1083/jcb.151.2.263.
- Ketelaar T, Voss C, Dimmock SA, Thumm M, Hussey PJ. Arabidopsis homologues of the autophagy protein Atg8 are a novel family of microtubule binding proteins. FEBS Lett. 2004;567(2–3):302–306. doi:https://doi.org/10.1016/j.febslet.2004.04.088.
- Zientara-Rytter K, Sirko A. Selective autophagy receptor Joka2 co-localizes with cytoskeleton in plant cells. Plant Signal Behav. 2014;9(3):e28523. doi:https://doi.org/10.4161/psb.28523.
- Wang Y, Zheng X, Yu B, Han S, Guo J, Tang H, Yu AY, Deng H, Hong Y, Liu Y. Disruption of microtubules in plants suppresses macroautophagy and triggers starch excess-associated chloroplast autophagy. Autophagy. 2015;11(12):2259–2274. doi:https://doi.org/10.1080/15548627.2015.1113365.
- Machesky LM, Insall RH. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8(25):1347–1356. doi:https://doi.org/10.1016/s0960-9822(98)00015-3.
- Pollard TD, Beltzner CC. Structure and function of the Arp2/3 complex. Curr Opin Struct Biol. 2002;12(6):768–774. doi:https://doi.org/10.1016/s0959-440x(02)00396-2.
- Deeks MJ, Hussey PJ. Arp2/3 and SCAR: plants move to the fore. Nat Rev Mol Cell Biol. 2005;6(12):954–964. doi:https://doi.org/10.1038/nrm1765.
- Szymanski D. Breaking the WAVE complex: the point of Arabidopsis trichomes. Curr Opin Plant Biol. 2005;8(1):103–112. doi:https://doi.org/10.1016/j.pbi.2004.11.004.
- Dyachok J, Shao MR, Vaughn K, Bowling A, Facette M, Djakovic S, Clark L, Smith L. Plasma membrane-associated SCAR complex subunits promote cortical F-actin accumulation and normal growth characteristics in Arabidopsis roots. Mol Plant. 2008;1(6):990–1006. doi:https://doi.org/10.1093/mp/ssn059.
- Wang P, Richardson C, Hawes C, Hussey PJ. Arabidopsis NAP1 regulates the formation of autophagosomes. Curr Biol. 2016;26(15):2060–2069. doi:https://doi.org/10.1016/j.cub.2016.06.008.
- Zheng X, Wu M, Li X, Cao J, Li J, Wang J, Huang S, Liu Y, Wang Y. Actin filaments are dispensable for bulk autophagy in plants. Autophagy. 2019;15(12):2126–2141. doi:https://doi.org/10.1080/15548627.2019.1596496.
- Lin Y, Guo R, Ji C, Zhou J, Jiang L. New insights into AtNBR1 as a selective autophagy cargo receptor in Arabidopsis. Plant Signal Behav. 2021;16(1):1839226. doi:https://doi.org/10.1080/15592324.2020.1839226.
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21(1):77–91. doi:https://doi.org/10.1016/j.devcel.2011.05.015.
- Winter V, Hauser MT. Exploring the ESCRTing machinery in eukaryotes. Trends Plant Sci. 2006;11(3):115–123. doi:https://doi.org/10.1016/j.tplants.2006.01.008.
- Gao C. Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc Natl Acad Sci U S A. 2015;112(112):1886–1891. doi:https://doi.org/10.1073/pnas.1421271112.
- Kolb C, Nagel MK, Kalinowska K, Hagmann J, Ichikawa M, Anzenberger F, Alkofer A, Sato MH, Braun P, Isono E. FYVE1 is essential for vacuole biogenesis and intracellular trafficking in Arabidopsis. Plant Physiol. 2015;167(4):1361–1373. doi:https://doi.org/10.1104/pp.114.253377.
- Katsiarimpa A, Kalinowska K, Anzenberger F, Weis C, Ostertag M, Tsutsumi C, Schwechheimer C, Brunner F, Huckelhoven R, Isono E. The deubiquitinating enzyme AMSH1 and the ESCRT-III subunit VPS2.1 are required for autophagic degradation in Arabidopsis. Plant Cell. 2013;25(6):2236–2252. doi:https://doi.org/10.1105/tpc.113.113399.
- Sutipatanasomboon A, Herberth S, Alwood EG, Haweker H, Muller B, Shahriari M, Zienert AY, Marin B, Robatzek S, Praefcke GJK, et al. Disruption of the plant-specific CFS1 gene impairs autophagosome turnover and triggers EDS1-dependent cell death. Sci Rep. 2017;7(1):8677. doi:https://doi.org/10.1038/s41598-017-08577-8.
- Isono E, Katsiarimpa A, Muller IK, Anzenberger F, Stierhof YD, Geldner N, Chory J, Schwechheimer C. The deubiquitinating enzyme AMSH3 is required for intracellular trafficking and vacuole biogenesis in Arabidopsis thaliana. Plant Cell. 2010;22(6):1826–1837. doi:https://doi.org/10.1105/tpc.110.075952.
- Kulich I, Pecenkova T, Sekeres J, Smetana O, Fendrych M, Foissner I, Hoftberger M, Zarsky V. Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic. 2013;14(11):1155–1165. doi:https://doi.org/10.1111/tra.12101.
- Surpin M, Zheng H, Morita MT, Saito C, Avila E, Blakeslee JJ, Bandyopadhyay A, Kovaleva V, Carter D, Murphy A, et al. The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell. 2003;15(12):2885–2899. doi:https://doi.org/10.1105/tpc.016121.
- Zouhar J, Rojo E, Bassham DC. AtVPS45 is a positive regulator of the SYP41/SYP61/VTI12 SNARE complex involved in trafficking of vacuolar cargo. Plant Physiol. 2009;149(4):1668–1678. doi:https://doi.org/10.1104/pp.108.134361.
- Hyttinen JM, Niittykoski M, Salminen A, Kaarniranta K. Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim Biophys Acta. 2013;1833(3):503–510. doi:https://doi.org/10.1016/j.bbamcr.2012.11.018.
- Gutierrez MG. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117(13):2687–2697. doi:https://doi.org/10.1242/jcs.01114.
- Balderhaar HJK, Arlt H, Ostrowicz C, Brocker C, Sundermann F, Brandt R, Babst M, Ungermann C. The Rab GTPase Ypt7 is linked to retromer-mediated receptor recycling and fusion at the yeast late endosome. J Cell Sci. 2010;123(23):4085–4094. doi:https://doi.org/10.1242/jcs.071977.
- Kwon SI, Cho HJ, Jung JH, Yoshimoto K, Shirasu K, Park OK. The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J. 2010;64(1):151–164. doi:https://doi.org/10.1111/j.1365-313X.2010.04315.x.
- Kwon SI, Cho HJ, Kim SR, Park OK. The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiol. 2013;161(4):1722–1736. doi:https://doi.org/10.1104/pp.112.208108.
- Zeng Y, Li B, Ji C, Feng L, Niu F, Deng C, Chen S, Lin Y, Cheung KCP, Shen J, et al. A unique AtSar1D-AtRabD2a nexus modulates autophagosome biogenesis in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2021;118(17):e2021293118. doi:https://doi.org/10.1073/pnas.2021293118.
- Zeng Y, Jiang L. A unique COPII population in plant autophagy. Autophagy. 2021:1–3. doi:https://doi.org/10.1080/15548627.2021.1933298.
- Yang X, Bassham DC. New insight into the mechanism and function of autophagy in plant cells. Int Rev Cell Mol Biol. 2015;320:1–40. doi:https://doi.org/10.1016/bs.ircmb.2015.07.005.
- Rojo E, Zouhar J, Carter C, Kovaleva V, Raikhel NV. A unique mechanism for protein processing and degradation in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2003;100(12):7389–7394. doi:https://doi.org/10.1073/pnas.1230987100.
- Sze H. Energization of plant cell membranes by H+-Pumping ATPases: regulation and biosynthesis. Plant Cell. 1999;11(4):677–689. doi:https://doi.org/10.1105/tpc.11.4.677.
- Kriegel A, Andres Z, Medzihradszky A, Kruger F, Scholl S, Delang S, Patir-Nebioglu MG, Gute G, Yang H, Murphy AS, et al. Job sharing in the endomembrane system: vacuolar acidification requires the combined activity of V-ATPase and V-PPase. Plant Cell. 2015;27(12):3383–3396. doi:https://doi.org/10.1105/tpc.15.00733.
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005;138(4):2097–2110. doi:https://doi.org/10.1104/pp.105.060673.
- Yu J, Zhou J. Vacuolar accumulation and colocalization is not a proper criterion for cytoplasmic soluble proteins undergoing selective autophagy. Plant Signal Behav. 2021;16(10):1932319. doi:https://doi.org/10.1080/15592324.2021.1932319.
- Slavikova S, Shy G, Yao Y, Glozman R, Levanony H, Pietrokovski S, Elazar Z, Galili G. The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J Exp Bot. 2005;56(421):2839–2849. doi:https://doi.org/10.1093/jxb/eri276.
- Inoue Y, Suzuki T, Hattori M, Yoshimoto K, Ohsumi Y, Moriyasu Y. AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 2006;47(12):1641–1652. doi:https://doi.org/10.1093/pcp/pcl031.
- Liu Y, Xiong Y, Bassham DC. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy. 2009;5(7):954–963. doi:https://doi.org/10.4161/auto.5.7.9290.
- Hayward AP, Dinesh-Kumar SP. What can plant autophagy do for an innate immune response? Annu Rev Phytopathol. 2011;49:557–576. doi:https://doi.org/10.1146/annurev-phyto-072910-095333.
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell. 2011;23(3):873–894. doi:https://doi.org/10.1105/tpc.111.083345.
- Ren C, Liu J, Gong Q. Functions of autophagy in plant carbon and nitrogen metabolism. Front Plant Sci. 2014;5:301. doi:https://doi.org/10.3389/fpls.2014.00301.
- Yang C, Luo M, Zhuang X, Li F, Gao C. Transcriptional and epigenetic regulation of autophagy in plants. Trends Genet. 2020;36(9):676–688. doi:https://doi.org/10.1016/j.tig.2020.06.013.
- Smith AM, Stitt M. Coordination of carbon supply and plant growth. Plant Cell Environ. 2007;30(9):1126–1149. doi:https://doi.org/10.1111/j.1365-3040.2007.01708.x.
- Wang Y, Yu B, Zhao J, Guo J, Li Y, Han S, Huang L, Du Y, Hong Y, Tang D, et al. Autophagy contributes to leaf starch degradation. Plant Cell. 2013;25(4):1383–1399. doi:https://doi.org/10.1105/tpc.112.108993.
- Izumi M, Hidema J, Makino A, Ishida H. Autophagy contributes to nighttime energy availability for growth in Arabidopsis. Plant Physiol. 2013;161(4):1682–1693. doi:https://doi.org/10.1104/pp.113.215632.
- Araujo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR. Protein degradation - an alternative respiratory substrate for stressed plants. Trends Plant Sci. 2011;16(9):489–498. doi:https://doi.org/10.1016/j.tplants.2011.05.008.
- Kurusu T, Koyano T, Hanamata S, Kubo T, Noguchi Y, Yagi C, Nagata N, Yamamoto T, Ohnishi T, Okazaki Y, et al. OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy. 2014;10(5):878–888. doi:https://doi.org/10.4161/auto.28279.
- Fan J, Yu L, Xu C. Dual role for autophagy in lipid metabolism in Arabidopsis. Plant Cell. 2019;31(7):1598–1613. doi:https://doi.org/10.1105/tpc.19.00170.
- Barros JAS, Magen S, Lapidot-Cohen T, Rosental L, Brotman Y, Araujo WL, Avin-Wittenberg T. Autophagy is required for lipid homeostasis during dark-induced senescence. Plant Physiol. 2021;185(4):1542–1558. doi:https://doi.org/10.1093/plphys/kiaa120.
- Izumi M, Hidema J, Ishida H. Deficiency of autophagy leads to significant changes of metabolic profiles in Arabidopsis. Plant Signal Behav. 2013;8(8):e25023. doi:https://doi.org/10.4161/psb.25023.
- Avin-Wittenberg T, Bajdzienko K, Wittenberg G, Alseekh S, Tohge T, Bock R, Giavalisco P, Fernie AR. Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cell. 2015;27(2):306–322. doi:https://doi.org/10.1105/tpc.114.134205.
- Thirumalaikumar VP, Wagner M, Balazadeh S, Skirycz A. Autophagy is responsible for the accumulation of proteogenic dipeptides in response to heat stress in Arabidopsis thaliana. FEBS J. 2021;288(1):281–292. doi:https://doi.org/10.1111/febs.15336.
- McLoughlin F, Augustine RC, Marshall RS, Li F, Kirkpatrick LD, Otegui MS, Vierstra RD. Maize multi-omics reveal roles for autophagic recycling in proteome remodelling and lipid turnover. Nat Plants. 2018;4(12):1056–1070. doi:https://doi.org/10.1038/s41477-018-0299-2.
- McLoughlin F, Marshall RS, Ding X, Chatt EC, Kirkpatrick LD, Augustine RC, Li F, Otegui MS, Vierstra RD. Autophagy plays prominent roles in amino acid, nucleotide, and carbohydrate metabolism during fixed-carbon starvation in maize. Plant Cell. 2020;32(9):2699–2724. doi:https://doi.org/10.1105/tpc.20.00226.
- Osmond MB. Effects of nitrogen nutrition on nitrogen partitioning between chloroplasts and mitochondria in pea and wheat. Plant Physiol. 1991;96(2):355–362. doi:https://doi.org/10.1104/pp.96.2.355.
- Guiboileau A, Yoshimoto K, Soulay F, Bataille MP, Avice JC, Masclaux-Daubresse C. Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 2012;194(3):732–740. doi:https://doi.org/10.1111/j.1469-8137.2012.04084.x.
- Guiboileau A, Avila-Ospina L, Yoshimoto K, Soulay F, Azzopardi M, Marmagne A, Lothier J, Masclaux-Daubresse C. Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytol. 2013;199(3):683–694. doi:https://doi.org/10.1111/nph.12307.
- Wada S, Hayashida Y, Izumi M, Kurusu T, Hanamata S, Kanno K, Kojima S, Yamaya T, Kuchitsu K, Makino A, et al. Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol. 2015;168(1):60–73. doi:https://doi.org/10.1104/pp.15.00242.
- Xia T, Xiao D, Liu D, Chai W, Gong Q, Wang NN. Heterologous expression of ATG8c from soybean confers tolerance to nitrogen deficiency and increases yield in Arabidopsis. PloS One. 2012;7(5):e37217. doi:https://doi.org/10.1371/journal.pone.0037217.
- Chen Q, Soulay F, Saudemont B, Elmayan T, Marmagne A, Masclaux-Daubresse CL. Overexpression of ATG8 in Arabidopsis stimulates autophagic activity and increases nitrogen remobilization efficiency and grain filling. Plant Cell Physiol. 2019;60(2):343–352. doi:https://doi.org/10.1093/pcp/pcy214.
- Phillips AR, Suttangkakul A, Vierstra RD. The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics. 2008;178(3):1339–1353. doi:https://doi.org/10.1534/genetics.107.086199.
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem. 2002;277(36):33105–33114. doi:https://doi.org/10.1074/jbc.M204630200.
- Masclaux-Daubresse C, Clement G, Anne P, Routaboul JM, Guiboileau A, Soulay F, Shirasu K, Yoshimoto K. Stitching together the multiple dimensions of autophagy using metabolomics and transcriptomics reveals impacts on metabolism, development, and plant responses to the environment in Arabidopsis. Plant Cell. 2014;26(5):1857–1877. doi:https://doi.org/10.1105/tpc.114.124677.
- Tegeder M, Rentsch D. Uptake and partitioning of amino acids and peptides. Mol Plant. 2010;3(6):997–1011. doi:https://doi.org/10.1093/mp/ssq047.
- Hillmer S, Movafeghi A, Robinson DG, Hinz G. Vacuolar storage proteins are sorted in the cis-cisternae of the pea cotyledon Golgi apparatus. J Cell Biol. 2001;152(1):41–50. doi:https://doi.org/10.1083/jcb.152.1.41.
- Ibl V, Stoger E. The formation, function and fate of protein storage compartments in seeds. Protoplasma. 2012;249(2):379–392. doi:https://doi.org/10.1007/s00709-011-0288-z.
- Chrispeels MJ, Herman EM. Endoplasmic reticulum-derived compartments function in storage and as mediators of vacuolar remodeling via a new type of organelle, precursor protease vesicles. Plant Physiol. 2000;123(4):1227–1233. doi:https://doi.org/10.1104/pp.123.4.1227.
- Bassham DC. Golgi-independent trafficking of macromolecules to the plant vacuole. Adv Bot Res. 2002;38:65–92. doi:https://doi.org/10.1016/S0065-2296(02)38028-5.
- Angelovici R, Fait A, Zhu X, Szymanski J, Feldmesser E, Fernie AR, Galili G. Deciphering transcriptional and metabolic networks associated with lysine metabolism during Arabidopsis seed development. Plant Physiol. 2009;151(4):2058–2072. doi:https://doi.org/10.1104/pp.109.145631.
- Di Berardino J, Marmagne A, Berger A, Yoshimoto K, Cueff G, Chardon F, Masclaux-Daubresse C, Reisdorf-Cren M. Autophagy controls resource allocation and protein storage accumulation in Arabidopsis seeds. J Exp Bot. 2018;69(6):1403–1414. doi:https://doi.org/10.1093/jxb/ery012.
- Chung T, Suttangkakul A, Vierstra RD. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 2009;149(1):220–234. doi:https://doi.org/10.1104/pp.108.126714.
- Li F, Chung T, Pennington JG, Federico ML, Kaeppler HF, Kaeppler SM, Otegui MS, Vierstra RD. Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell. 2015;27(5):1389–1408. doi:https://doi.org/10.1105/tpc.15.00158.
- Honig A, Avin-Wittenberg T, Ufaz S, Galili G. A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell. 2012;24(1):288–303. doi:https://doi.org/10.1105/tpc.111.093112.
- Xiong Y, Contento AL, Bassham DC. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005;42(4):535–546. doi:https://doi.org/10.1111/j.1365-313X.2005.02397.x.
- Lee TA, Vande Wetering SW, Brusslan JA. Stromal protein degradation is incomplete in Arabidopsis thaliana autophagy mutants undergoing natural senescence. BMC Res Notes. 2013;6:17. doi:https://doi.org/10.1186/1756-0500-6-17.
- Kwon SI, Cho HJ, Park OK. Role of Arabidopsis RabG3b and autophagy in tracheary element differentiation. Autophagy. 2010;6(8):1187–1189. doi:https://doi.org/10.4161/auto.6.8.13429.
- Huang L, Yu LJ, Zhang X, Fan B, Wang FZ, Dai YS, Qi H, Zhou Y, Xie LJ, Xiao S. Autophagy regulates glucose-mediated root meristem activity by modulating ROS production in Arabidopsis. Autophagy. 2019;15(3):407–422. doi:https://doi.org/10.1080/15548627.2018.1520547.
- Zhang Y, Li S, Zhou LZ, Fox E, Pao J, Sun W, Zhou C, McCormick S. Overexpression of Arabidopsis thaliana PTEN caused accumulation of autophagic bodies in pollen tubes by disrupting phosphatidylinositol 3-phosphate dynamics. Plant J. 2011;68(6):1081–1092. doi:https://doi.org/10.1111/j.1365-313X.2011.04761.x.
- Fujiki Y, Yoshimoto K, Ohsumi Y. An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 2007;143(3):1132–1139. doi:https://doi.org/10.1104/pp.106.093864.
- Harrison-Lowe NJ, Olsen LJ. Autophagy protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. Autophagy. 2008;4(3):339–348. doi:https://doi.org/10.4161/auto.5629.
- Wang WY, Zhang L, Xing S, Ma Z, Liu J, Gu H, Qin G, Qu LJ. Arabidopsis AtVPS15 plays essential roles in pollen germination possibly by interacting with AtVPS34. J Genet Genomics. 2012;39(2):81–92. doi:https://doi.org/10.1016/j.jgg.2012.01.002.
- Qin G, Ma Z, Zhang L, Xing S, Hou X, Deng J, Liu J, Chen Z, Qu LJ, Gu H. Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res. 2007;17(3):249–263. doi:https://doi.org/10.1038/cr.2007.7.
- Hanamata S, Sawada J, Ono S, Ogawa K, Fukunaga T, Nonomura KI, Kimura S, Kurusu T, Kuchitsu K. Impact of autophagy on gene expression and tapetal programmed cell death during pollen development in rice. Front Plant Sci. 2020;11:172. doi:https://doi.org/10.3389/fpls.2020.00172.
- Dundar G, Shao Z, Higashitani N, Kikuta M, Izumi M, Higashitani A. Autophagy mitigates high-temperature injury in pollen development of Arabidopsis thaliana. Dev Biol. 2019;456(2):190–200. doi:https://doi.org/10.1016/j.ydbio.2019.08.018.
- Zhao P, Zhou XM, Zhao LL, Cheung AY, Sun MX. Autophagy-mediated compartmental cytoplasmic deletion is essential for tobacco pollen germination and male fertility. Autophagy. 2020;16(12):2180–2192. doi:https://doi.org/10.1080/15548627.2020.1719722.
- Hanamata S, Kurusu T, Kuchitsu K. Roles of autophagy in male reproductive development in plants. Front Plant Sci. 2014;5:457. doi:https://doi.org/10.3389/fpls.2014.00457.
- Norizuki T, Minamino N, Ueda T. Role of autophagy in male reproductive processes in land plants. Front Plant Sci. 2020;11:756. doi:https://doi.org/10.3389/fpls.2020.00756.
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi:https://doi.org/10.1038/nature05286.
- Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat Rev Genet. 2010;11(8):539–548. doi:https://doi.org/10.1038/nrg2812.
- Liu Y, Bassham DC. Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol. 2012;63:215–237. doi:https://doi.org/10.1146/annurev-arplant-042811-105441.
- Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121(4):567–577. doi:https://doi.org/10.1016/j.cell.2005.03.007.
- Patel S, Dinesh-Kumar SP. Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy. 2008;4(1):20–27. doi:https://doi.org/10.4161/auto.5056.
- Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, Jorgensen LB, Jones JD, Mundy J, Petersen M. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009;137(4):773–783. doi:https://doi.org/10.1016/j.cell.2009.02.036.
- Hofius D, Mundy J, Petersen M. Self-consuming innate immunity in Arabidopsis. Autophagy. 2009;5(8):1206–1207. doi:https://doi.org/10.4161/auto.5.8.9892.
- Lenz HD, Haller E, Melzer E, Kober K, Wurster K, Stahl M, Bassham DC, Vierstra RD, Parker JE, Bautor J, et al. Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 2011;66(5):818–830. doi:https://doi.org/10.1111/j.1365-313X.2011.04546.x.
- Lenz HD, Vierstra RD, Nürnberger T, Gust AA. ATG7 contributes to plant basal immunity towards fungal infection. Plant Signal Behav. 2011;6(7):1040–1042. doi:https://doi.org/10.4161/psb.6.7.15605.
- Li Y, Kabbage M, Liu W, Dickman MB. Aspartyl protease-mediated cleavage of BAG6 is necessary for autophagy and fungal resistance in plants. Plant Cell. 2016;28(1):233–247. doi:https://doi.org/10.1105/tpc.15.00626.
- Henry E, Fung N, Liu J, Drakakaki G, Coaker G. Beyond glycolysis: gAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genet. 2015;11(4):e1005199. doi:https://doi.org/10.1371/journal.pgen.1005199.
- Han S, Wang Y, Zheng X, Jia Q, Zhao J, Bai F, Hong Y, Liu Y. Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell. 2015;27(4):1316–1331. doi:https://doi.org/10.1105/tpc.114.134692.
- Zhang B, Shao L, Wang J, Zhang Y, Guo X, Peng Y, Cao Y, Lai Z. Phosphorylation of ATG18a by BAK1 suppresses autophagy and attenuates plant resistance against necrotrophic pathogens. Autophagy. 2020;1–18. doi:https://doi.org/10.1080/15548627.2020.1810426.
- Brillada C, Teh OK, Ditengou FA, Lee CW, Klecker T, Saeed B, Furlan G, Zietz M, Hause G, Eschen-Lippold L, et al. Exocyst subunit Exo70B2 is linked to immune signaling and autophagy. Plant Cell. 2021;33(2):404–419. doi:https://doi.org/10.1093/plcell/koaa022.
- Koornneef A, Pieterse CM. Cross talk in defense signaling. Plant Physiol. 2008;146(3):839–844. doi:https://doi.org/10.1104/pp.107.112029.
- Spoel SH, Dong X. Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe. 2008;3(6):348–351. doi:https://doi.org/10.1016/j.chom.2008.05.009.
- De Vleesschauwer D, Xu J, Hofte M. Making sense of hormone-mediated defense networking: from rice to Arabidopsis. Front Plant Sci. 2014;5:611. doi:https://doi.org/10.3389/fpls.2014.00611.
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K. Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell. 2009;21(9):2914–2927. doi:https://doi.org/10.1105/tpc.109.068635.
- Wang Y, Nishimura MT, Zhao T, Tang D. ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 2011;68(1):74–87. doi:https://doi.org/10.1111/j.1365-313X.2011.04669.x.
- Hofius D, Munch D, Bressendorff S, Mundy J, Petersen M. Role of autophagy in disease resistance and hypersensitive response-associated cell death. Cell Death Differ. 2011;18(8):1257–1262. doi:https://doi.org/10.1038/cdd.2011.43.
- Kim SH, Kwon C, Lee JH, Chung T. Genes for plant autophagy: functions and interactions. Mol Cells. 2012;34(5):413–423. doi:https://doi.org/10.1007/s10059-012-0098-y.
- Lal NK, Thanasuwat B, Chan B, Dinesh-Kumar SP. Pathogens manipulate host autophagy through injected effector proteins. Autophagy. 2020;16(12):2301–2302. doi:https://doi.org/10.1080/15548627.2020.1831816.
- Lal NK, Thanasuwat B, Huang P-j, Cavanaugh KA, Carter A, Michelmore RW, Dinesh-Kumar SP. Phytopathogen effectors use multiple mechanisms to manipulate plant autophagy. Cell Host Microbe. 2020;28(4):558–571.e556. doi:https://doi.org/10.1016/j.chom.2020.07.010.
- Dagdas YF, Khaoula B, Abbas M, Angela CG, Pooja P, Benjamin P, Nadra T, Neftaly CM, Hughes RK, Jan S. An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. eLife. 2016;14(5):e10856. doi:https://doi.org/10.7554/eLife.10856.
- Maqbool A, Hughes RK, Dagdas YF, Tregidgo N, Zess E, Belhaj K, Round A, Bozkurt TO, Kamoun S, Banfield MJ. Structural basis of host autophagy-related protein 8 (ATG8) binding by the Irish potato famine pathogen effector protein PexRD54. J Biol Chem. 2016;291(38):20270–20282. doi:https://doi.org/10.1074/jbc.M116.744995.
- Yang M, Ismayil A, Liu Y. Autophagy in plant-virus interactions. Annual Review of Virology. 2020;7(1):1–17. doi:https://doi.org/10.1146/annurev-virology-010220-054709.
- Choi Y, Bowman JW, Jung JU. Autophagy during viral infection - a double-edged sword. Nat Rev Microbiol. 2018;16(6):341–354. doi:https://doi.org/10.1038/s41579-018-0003-6.
- Gorovits R, Moshe A, Ghanim M, Czosnek H. Degradation mechanisms of the Tomato yellow leaf curl virus coat protein following inoculation of tomato plants by the whitefly Bemisia tabaci. Pest Manag Sci. 2014;70(10):1632–1639. doi:https://doi.org/10.1002/ps.3737.
- Gorovits R, Fridman L, Kolot M, Rotem O, Ghanim M, Shriki O, Czosnek H. Tomato yellow leaf curl virus confronts host degradation by sheltering in small/midsized protein aggregates. Virus Res. 2016;213:304–313. doi:https://doi.org/10.1016/j.virusres.2015.11.020.
- Haxim Y, Ismayil A, Qi J, Yan W, Liu Y. Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife. 2017;6:e23897. doi:https://doi.org/10.7554/eLife.23897.
- Hafrén A, Macia JL, Love AJ, Milner JJ, Drucker M, Hofius D. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc Natl Acad Sci U S A. 2017;114(10):E2026–E2035. doi:https://doi.org/10.1073/pnas.1610687114.
- Li F, Zhang M, Zhang C, Zhou X. Nuclear autophagy degrades a geminivirus nuclear protein to restrict viral infection in solanaceous plants. New Phytol. 2020;225(4):1746–1761. doi:https://doi.org/10.1111/nph.16268.
- Yang M, Zhang Y, Xie X, Yue N, Li J, Wang XB, Han C, Yu J, Liu Y, Li D. Barley stripe mosaic virus γb protein subverts autophagy to promote viral infection by disrupting the ATG7-ATG8 interaction. Plant Cell. 2018;30(7):1582–1595. doi:https://doi.org/10.1105/tpc.18.00122.
- Hafren A, Ustun S, Hochmuth A, Svenning S, Johansen T, Hofius D. Turnip mosaic virus counteracts selective autophagy of the viral silencing suppressor HCpro. Plant Physiol. 2018;176(1):649–662. doi:https://doi.org/10.1104/pp.17.01198.
- Li F, Zhang C, Li Y, Wu G, Hou X, Zhou X, Wang A. Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral polymerase. Nat Commun. 2018;9(1):1268. doi:https://doi.org/10.1038/s41467-018-03658-2.
- Fu S, Xu Y, Li C, Li Y, Wu J, Zhou X. Rice stripe virus interferes with S-acylation of remorin and induces its autophagic degradation to facilitate virus infection. Mol Plant. 2018;11(2):269–287. doi:https://doi.org/10.1016/j.molp.2017.11.011.
- Xiong Y, Contento AL, Nguyen PQ, Bassham DC. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007;143(1):291–299. doi:https://doi.org/10.1104/pp.106.092106.
- Zhou J, Wang J, Cheng Y, Chi YJ, Fan B, Yu JQ, Chen Z. NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PLoS Genet. 2013;9(1):e1003196. doi:https://doi.org/10.1371/journal.pgen.1003196.
- Gassmann W, Zhou J, Zhang Y, Qi J, Chi Y, Fan B, Yu JQ, Chen Z. E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 2014;10(1):e1004116. doi:https://doi.org/10.1371/journal.pgen.1004116.
- Kuzuoglu-Ozturk D, Cebeci Yalcinkaya O, Akpinar BA, Mitou G, Korkmaz G, Gozuacik D, Budak H. Autophagy-related gene, TdAtg8, in wild emmer wheat plays a role in drought and osmotic stress response. Planta. 2012;236(4):1081–1092. doi:https://doi.org/10.1007/s00425-012-1657-3.
- Pei D, Zhang W, Sun H, Wei X, Yue J, Wang H. Identification of autophagy-related genes ATG4 and ATG8 from wheat (Triticum aestivum L.) and profiling of their expression patterns responding to biotic and abiotic stresses. Plant Cell Rep. 2014;33(10):1697–1710. doi:https://doi.org/10.1007/s00299-014-1648-x.
- Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Arch Biochem Biophys. 2005;444(2):139–158. doi:https://doi.org/10.1016/j.abb.2005.10.018.
- Leshem Y, Seri L, Levine A. Induction of phosphatidylinositol 3-kinase-mediated endocytosis by salt stress leads to intracellular production of reactive oxygen species and salt tolerance. Plant J. 2007;51(2):185–197. doi:https://doi.org/10.1111/j.1365-313X.2007.03134.x.
- Slavikova S, Ufaz S, Avin-Wittenberg T, Levanony H, Galili G. An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J Exp Bot. 2008;59(14):4029–4043. doi:https://doi.org/10.1093/jxb/ern244.
- Shin JH, Yoshimoto K, Ohsumi Y, Jeon JS, An G. OsATG10b, an autophagosome component, is needed for cell survival against oxidative stresses in rice. Mol Cells. 2009;27(1):67–74. doi:https://doi.org/10.1007/s10059-009-0006-2.
- Luo L, Zhang P, Zhu R, Fu J, Su J, Zheng J, Wang Z, Wang D, Gong Q. Autophagy is rapidly induced by salt stress and is required for salt tolerance in Arabidopsis. Front Plant Sci. 2017;8:1459. doi:https://doi.org/10.3389/fpls.2017.01459.
- Sung DY, Kaplan F, Lee KJ, Guy CL. Acquired tolerance to temperature extremes. Trends Plant Sci. 2003;8(4):179–187. doi:https://doi.org/10.1016/s1360-1385(.03)00047-5.
- Zhou J, Wang J, Yu JQ, Chen Z. Role and regulation of autophagy in heat stress responses of tomato plants. Front Plant Sci. 2014;5:174. doi:https://doi.org/10.3389/fpls.2014.00174.
- Sedaghatmehr M, Thirumalaikumar VP, Kamranfar I, Marmagne A, Masclaux-Daubresse C, Balazadeh S. A regulatory role of autophagy for resetting the memory of heat stress in plants. Plant Cell Environ. 2019;42(3):1054–1064. doi:https://doi.org/10.1111/pce.13426.
- Jiang Z, Zhu L, Wang Q, Hou X. Autophagy-related 2 regulates chlorophyll degradation under abiotic stress conditions in Arabidopsis. Int J Mol Sci. 2020;21(12):4515. doi:https://doi.org/10.3390/ijms21124515.
- Zhang Y, Min H, Shi C, Xia G, Lai Z. Transcriptome analysis of the role of autophagy in plant response to heat stress. PLoS One. 2021;16(2):e0247783. doi:https://doi.org/10.1371/journal.pone.0247783.
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi:https://doi.org/10.1038/nrm2199.
- Minina EA, Moschou PN, Vetukuri RR, Sanchez-Vera V, Cardoso C, Liu Q, Elander PH, Dalman K, Beganovic M, Lindberg Yilmaz J, et al. Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J Exp Bot. 2018;69(6):1415–1432. doi:https://doi.org/10.1093/jxb/ery010.
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40(2):280–293. doi:https://doi.org/10.1016/j.molcel.2010.09.023.
- Chen L, Liao B, Qi H, Xie LJ, Huang L, Tan WJ, Zhai N, Yuan LB, Zhou Y, Yu LJ, et al. Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy. 2015;11(12):2233–2246. doi:https://doi.org/10.1080/15548627.2015.1112483.
- Yamauchi S, Mano S, Oikawa K, Hikino K, Teshima KM, Kimori Y, Nishimura M, Shimazaki KI, Takemiya A. Autophagy controls reactive oxygen species homeostasis in guard cells that is essential for stomatal opening. Proc Natl Acad Sci U S A. 2019;116(38):19187–19192. doi:https://doi.org/10.1073/pnas.1910886116.
- Luo S, Li X, Zhang Y, Fu Y, Fan B, Zhu C, Chen Z. Cargo recognition and function of selective autophagy receptors in plants. Int J Mol Sci. 2021;22(3):1013. doi:https://doi.org/10.3390/ijms22031013.
- Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584(7):1379–1385. doi:https://doi.org/10.1016/j.febslet.2010.01.018.
- Marshall RS, Li F, Gemperline DC, Book AJ, Vierstra RD. Autophagic degradation of the 26s proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol Cell. 2015;58(6):1053–1066. doi:https://doi.org/10.1016/j.molcel.2015.04.023.
- Marshall RS, Hua Z, Mali S, McLoughlin F, Vierstra RD. ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell. 2019;177(3):766–781. doi:https://doi.org/10.1016/j.cell.2019.02.009.
- Li X, Liu Q, Feng H, Deng J, Zhang R, Wen J, Dong J, Wang T. Dehydrin MtCAS31 promotes autophagic degradation under drought stress. Autophagy. 2019;16(5):862–877. doi:https://doi.org/10.1080/15548627.2019.1643656.
- Bao Y, Song WM, Wang P, Yu X, Bassham DC. COST1 regulates autophagy to control plant drought tolerance. Proc Natl Acad Sci U S A. 2020;117(13):7482–7493. doi:https://doi.org/10.1073/pnas.1918539117.
- Nolan TM, Brennan B, Yang M, Chen J, Zhang M, Li Z, Wang X, Bassham DC, Walley J, Yin Y. Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev Cell. 2017;41(1):33–46. doi:https://doi.org/10.1016/j.devcel.2017.03.013.
- Guillaumot D, Guillon S, Deplanque T, Vanhee C, Gumy C, Masquelier D, Morsomme P, Batoko H. The Arabidopsis TSPO-related protein is a stress and abscisic acid-regulated, endoplasmic reticulum-Golgi-localized membrane protein. Plant J. 2009;60(2):242–256. doi:https://doi.org/10.1111/j.1365-313X.2009.03950.x.
- Balsemão-Pires E, Jaillais Y, Olson BJ, Andrade LR, Umen JG, Chory J, Sachetto-Martins G. The Arabidopsis translocator protein (AtTSPO) is regulated at multiple levels in response to salt stress and perturbations in tetrapyrrole metabolism. BMC Plant Biol. 2011;11(1):1–17. doi:https://doi.org/10.1186/1471-2229-11-108.
- Vanhee C, Zapotoczny G, Masquelier D, Ghislain M, Batoko H. The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell. 2011;23(2):785–805. doi:https://doi.org/10.1105/tpc.110.081570.
- Michaeli S, Honig A, Levanony H, Peled-Zehavi H, Galili G. Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell. 2014;26(10):4084–4101. doi:https://doi.org/10.1105/tpc.114.129999.
- Zhou J, Wang Z, Wang X, Li X, Zhang Z, Fan B, Zhu C, Chen Z. Dicot-specific ATG8-interacting ATI3 proteins interact with conserved UBAC2 proteins and play critical roles in plant stress responses. Autophagy. 2018;14(3):487–504. doi:https://doi.org/10.1080/15548627.2017.1422856.
- Thirumalaikumar VP, Gorka M, Schulz K, Masclaux-Daubresse C, Sampathkumar A, Skirycz A, Vierstra RD, Balazadeh S. Selective autophagy regulates heat stress memory in Arabidopsis by NBR1-mediated targeting of HSP90 and ROF1. Autophagy. 2020;1–16. doi:https://doi.org/10.1080/15548627.2020.1820778. Online ahead of print.
- Chi C, Li X, Fang P, Xia X, Shi K, Zhou Y, Zhou J, Yu J. Brassinosteroids act as a positive regulator of NBR1-dependent selective autophagy in response to chilling stress in tomato. J Exp Bot. 2020;71(3):1092–1106. doi:https://doi.org/10.1093/jxb/erz466.
- Su W, Bao Y, Lu Y, He F, Liu C. Poplar autophagy receptor NBR1 enhances salt stress tolerance by regulating selective autophagy and antioxidant system. Front Plant Sci. 2021;11:568411. doi:https://doi.org/10.3389/fpls.2020.568411.
- Zhang Y, Chen Z. Broad and complex roles of NBR1-mediated selective autophagy in plant stress responses. Cells. 2020;9(12):2562. doi:https://doi.org/10.3390/cells9122562.
- Luo S, Li X, Zhang Y, Fu Y, Fan B, Zhu C, Chen Z. Cargo recognition and function of selective autophagy receptors in plants. Int J Mol Sci. 2021;22(3):1013. doi:https://doi.org/10.3390/ijms22031013.
- Qi H, Xia FN, Xiao S. Autophagy in plants: physiological roles and post-translational regulation. J Integr Plant Biol. 2021;63(1):161–179. doi:https://doi.org/10.1111/jipb.12941.
