ABSTRACT
Nonhost plants effectively block a vast number of nonadapted fungal pathogens at the preinvasive stage. On the host plants, adapted fungal pathogens such as Colletotrichum species invade into plant epidermal cell by penetration peg developed from melanized appressorium, followed by invasive hyphal extension. I reported nonadapted Colletotrichum fungi that showed an increased rate of melanized appressorium-mediated entry (MAE) into the pen2 mutant of nonhost Arabidopsis thaliana (hereafter Arabidopsis). It was also found that other MAE-type nonadapted Colletotrichum fungi with no penetration into the pen2 mutant invaded Arabidopsis in the presence of additional mutations such as edr1, gsh1, eds5, cas, and chup1 in the pen2 background. Thus, many immune components contribute to the preinvasive nonhost resistance (NHR) of Arabidopsis against Colletotrichum MAE, and PEN2-related defense takes priority over other defense pathways. Here, I show that among the above nonadapted fungi, Colletotrichum nymphaeae PL1-1-b exhibited relatively lower incompatibility with the nonhost Arabidopsis with increased MAE in each single mutant of edr1, gsh1, eds5, and cas, although other nonadapted fungi almost never invaded these single mutants. Based on the relationships between Colletotrichum MAE and the Arabidopsis immune-related components, Colletotrichum-Arabidopsis incompatibility and multilayered immunity in the preinvasive NHR of Arabidopsis are discussed in this study.
Fungal pathogens directly invade the plant epidermis, followed by the development of invasive hyphae inside; therefore, penetration into epidermal cells is the most crucial step for successful infection. Only a few adapted fungal pathogens can infect a specific plant (defined as the host). If the plant is a nonhost, a robust and broad-spectrum defense termed nonhost resistance (NHR) effectively prevents the invasion of a vast number of nonadapted fungi in incompatible interactions.Citation1,Citation2
Incompatible interactions between the model brassicaceous plant Arabidopsis thaliana (hereafter Arabidopsis) and nonadapted powdery mildew fungi, such as Blumeria graminis f. sp. hordei, have been well studied for NHR. Publications reported that pen1, pen2, pen3, pmr4, ataf1, pldδ, cam7, cyp81F2, and vps9a mutant plants allowed increased entry of these obligate biotrophs into the epidermis.Citation3–11 PENETRATION 1 (PEN1) syntaxin mediates a secretory pathway to the fungal penetration site.Citation3,Citation12 PENETRATION 2 (PEN2) myrosinase, CYTOCHROME P450 FAMILY 81 SUBFAMILY F POLYPEPTIDE 2 (CYP81F2) monooxygenase, and PENETRATION 3 (PEN3) transporter are involved in metabolism and transport of tryptophan-derived secondary metabolites,Citation5,Citation6,Citation8,Citation13,Citation14 while CALMODULIN 7 (CaM7) is a Ca2+ sensor which regulates PEN3 function.Citation10 POWDERY MILDEW RESISTANT 4 (PMR4) callose synthase functions in wound and papillary callose formation.Citation4 ARABIDOPSIS THALIANA ACTIVATING FACTOR 1 (ATAF1) transcription factor suppresses abscisic acid biosynthesis and signaling.Citation7 PHOSPHOLIPASE Dδ (PLDδ) catalyzes phosphatidic acid production on the membrane in stress responses.Citation9 VACUOLAR PROTEIN SORTING 9a (VPS9a) guanine-nucleotide exchange factor is required for the delivery of membrane material to the haustorial encasement.Citation11 Thus, many pathways and components including as-yet-unknown factors elaborately underpin the Arabidopsis NHR against powdery mildew fungi.
The hemibiotroph Colletotrichum fungus was also instrumental in the research on NHR in Arabidopsis. Of those mentioned above, pen1, pen2, pen3, pmr4, and cyp81F2 mutants were used for the fungal entry test with nonadapted Colletotrichum tropicale S9275, a mulberry isolate, and the contribution of PEN2, PEN3, and CYP81F2 to preinvasive defense were demonstrated.Citation15,Citation16 Hiruma et al. also performed screening of Arabidopsis mutants, and ENHANCED DISEASE RESISTANCE 1 (EDR1) and γ-GLUTAMYLCYSTEINE SYNTHETASE 1 (GSH1) were found as NHR contributors against entry of C. tropicale S9275 to the epidermis.Citation16,Citation17 EDR1 protein kinase mediates the induction of antifungal plant defensins.Citation16,Citation18 GSH1 is involved in the generation of PEN2-related metabolites through glutathione biosynthesis, while GSH1 is also required for postinvasive defense.Citation13,Citation19 Immune kinases BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED RECEPTOR KINASE 1 (BAK1), BOTRYTIS-INDUCED KINASE 1 (BIK1), and AVRPPHB SUSCEPTIBLE1 (PBS1)-LIKE 1 (PBL1) working in the defense signaling triggered by pathogen-associated molecular patterns were also involved in preinvasive NHR against C. tropicale S9275.Citation20 On all these mutant plants, however, C. tropicale S9275 could barely develop invasive hyphae through melanized appressorium, a typical infection structure of Colletotrichum fungi. Instead, this pathogen successfully entered the Arabidopsis epidermis when the carbohydrate-induced hyphal tip-based entry (HTE).Citation15–17,Citation20 HTE, only reported in C. tropicale S9275 at present, was not associated with appressorium formation, thereby suggesting an atypical infection mode with different morphogenesis after perception of carbohydrate nutrients released from the wounded sites of the plants.Citation15 Therefore, there is little knowledge on Arabidopsis NHR against melanized appressorium-mediated entry (MAE) into the epidermis by Colletotrichum fungi. This is in contrast to those against hemibiotroph Pyricularia oryzae (Syn. Magnaporthe oryzae), which also shows MAE during plant infection.Citation21–26 In P. oryzae-Arabidopsis incompatible interaction, PEN2 functions as a NHR contributor against MAE and P. oryzae slightly invaded pen2 mutant.Citation21 Furthermore, many other immune components such as ARABIDOPSIS G-PROTEIN β-SUBUNIT 1 (AGB1), POWDERY MILDEW RESISTANT 5 (PMR5), MILDEW RESISTANCE LOCUS O 2 (MLO2), MAP KINASE 6 (MPK6), ERECTA, SUPPRESSOR of BAK1-INTERACTING RECEPTOR-LIKE KINASE 1 (SOBIR1), and EXTRA-LARGE G PROTEIN 2 (XLG2), the single mutation of which did not affect the breakdown of penetration resistance, showed decreased preinvasive NHR in the pen2 background.Citation21–26
I have recently reported three nonadapted Colletotrichum fungi, C. nymphaeae PL1-1-b (MAFF240037), C. fioriniae CC1 (MAFF306550), and C. siamense MAF1 (MAFF243010), isolates from Japanese flowering cherry, cosmos, and apple, respectively, which all showed increased MAE into the pen2 mutant.Citation27 It was also found that two nonadapted Colletotrichum fungi, C. siamense COC4 (MAFF243696) and C. orbiculare 104-T (MAFF240422), isolates from hau tree and cucumber, respectively, could not invade pen2 single mutant, but was able to invade on pen2 mutant with additional mutations such as edr1, gsh1, eds5, cas, and chup1.Citation27 The MAE rates of C. fioriniae CC1 and C. siamense MAF1 into pen2 mutants were also increased in the presence of edr1, gsh1, eds5, cas, and chup1.Citation27 ENHANCED DISEASE SUSCEPTIBILITY 5 (EDS5) is a MATE family transporter that is required for the transport of the salicylic acid precursor outside the chloroplasts.Citation28,Citation29 CALCIUM-SENSING RECEPTOR (CAS) is a Ca2+-sensing receptor involved in transient Ca2+ signaling in chloroplasts during plant immunity.Citation30 CHLOROPLAST UNUSUAL POSITIONING 1 (CHUP1) is a regulator of epidermal chloroplast response, a newly discovered immune-related response.Citation27 These findings reveal that PEN2, EDR1, GSH1, EDS5, CAS, and CHUP1 all contribute to preinvasive NHR of Arabidopsis against Colletotrichum MAE. It is noteworthy that the potential effects of edr1, gsh1, eds5, cas, and chup1 mutations on the MAE of C. fioriniae CC1 and C. siamense MAF1 were actualized on the pen2 background.Citation27 Furthermore, quantitative real-time PCR analysis showed that many defense-related genes were induced after inoculation of C. fioriniae CC1 only in pen2 among all tested single mutants.Citation27 These results indicate that the PEN2-related pathway functions as a higher-layer preinvasive defense and takes priority over other defense pathways, at least in preinvasive NHR against these nonadapted MAE-type Colletotrichum fungi.
Intriguingly, among the above MAE-type nonadapted fungi (), C. nymphaeae PL1-1-b could partly achieve MAE in the epidermis of wild-type Arabidopsis Col-0, and the chup1 single mutant permitted an increase in the trend of the MAE rateCitation27 (). These results suggest the potential ability of C. nymphaeae PL1-1-b to overcome the higher-layer preinvasive defense(s) of Arabidopsis to a certain degree, although the PEN2-related defense pathway is still working against this pathogenCitation27 (). To clarify this point, the MAE rate of C. nymphaeae PL1-1-b into edr1, gsh1, eds5, and cas single mutants was quantified, and it was found that this pathogen showed a significant increase or increasing trend in the MAE rate in all single mutants compared to that in Col-0 (). These results indicate that C. nymphaeae PL1-1-b partly overcomes the higher-layer preinvasive defense(s) in Arabidopsis and that each immune pathway related to EDR1, GSH1, EDS5, or CAS is effective against this pathogen in the wild-type plant. Thus, C. nymphaeae PL1-1-b is not adapted to Arabidopsis, but its incompatibility with the nonhost Arabidopsis is relatively low.
Figure 1. Nonadapted C. nymphaeae PL1-1-b exhibited relatively lower incompatibility with the nonhost Arabidopsis and showed increased MAE into edr1, gsh1, eds5, and cas single mutants. (a) Pathogenicity of Colletotrichum fungi on wild-type Arabidopsis (Col-0). A conidial suspension of adapted C. higginsianum Abr1-5 and nonadapted C. nymphaeae PL1-1-b, C. fioriniae CC1, C. siamense MAF1, COC4, and C. orbiculare 104-T was inoculated onto cotyledons of Col-0 and incubated. The photographs were taken at 5 and 9 days post-inoculation (dpi). (b) Invasion of C. nymphaeae PL1-1-b into epidermis of Col-0 at 4 dpi. The white arrowhead and arrows indicate melanized appressorium and invasive hyphae, respectively. The black arrowhead represents melanized appressorium with no invasive hypha. Scale bar, 10 µm. (c) MAE rate of C. nymphaeae PL1-1-b into Arabidopsis mutants at 4 dpi. For quantification of MAE, 300 melanized appressoria were investigated. The means and SE were calculated from six independent plants. The asterisks indicate significant difference from the control (Col-0) (*P < .05, **P < .01, one-way ANOVA with Dunnett’s test).
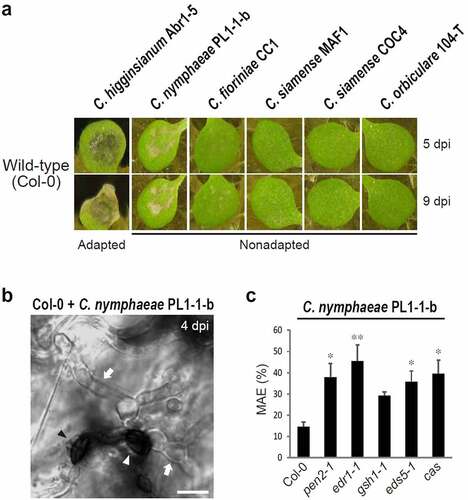
To visualize the relationships between Colletotrichum-Arabidopsis incompatibility and immune components in Arabidopsis preinvasive NHR, the Colletotrichum MAE rates were integrated from a previous studyCitation27 and newly obtained data, although I did not quantified the entry rates of C. nymphaeae PL1-1-b on Arabidopsis with multiple mutations, because the effect of each mutation was already actualized in the corresponding single mutants (). Consistent with the results of the pathogenicity assay of Colletotrichum fungi on the wild-type Arabidopsis Col-0 (), the MAE rates of five nonadapted fungi were notably lower than those of the adapted fungus C. higginsianum Abr1-5 (MAFF305635), a Brassicaceae pathogen (). Five nonadapted Colletotrichum strains displayed differential ability to overcome preinvasive NHR in Arabidopsis mutants in the following order: C. nymphaeae PL1-1-b > C. fioriniae CC1 > C. siamense MAF1 > C. siamense COC4 > C. orbiculare 104-T. C. nymphaeae, C. fioriniae, and C. siamense are classified into Colletotrichum clades with broad host range (acutatum or gloeosporioides). Thus, these nonadapted strains might exhibit lower incompatibility compared to C. orbiculare 104-T which generally shows narrow host range. Since I used one representative strain from one species, except C. siamense, it is difficult to confirm that the Arabidopsis response is specific to the fungal species. On the other hand, the interaction of each nonadapted strain with the nonhost Arabidopsis provided a clear insight into preinvasive NHR in Arabidopsis. To discuss the general interactions between fungal species and Arabidopsis, it is important to evaluate the interaction of multiple fungal strains from one species with Arabidopsis.
Figure 2. Relationships between MAE of nonadapted Colletotrichum fungi and preinvasive NHR in Arabodipsis thaliana. Invasion abilities of nonadapted C. nymphaeae PL1-1-b, C. fioriniae CC1, C. siamense MAF1, COC4, and C. orbiculare 104-T into nonhost Arabidopsis mutants were evaluated based on the MAE rates and classified. Adapted C. higginsianum Abr1-5 was showed as a control. PEN2-related defense takes priority over other immune components. The percentage of fungal entry test as “-”, “+/-”, “+”, “++”, “+++”, and “++++” was 0–2%, 2–10%, 10–20%, 20–35%, 35–60%, and 60–100%, respectively. n.d.: not determined.
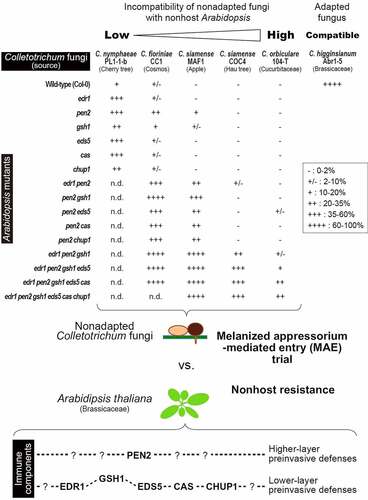
Apart from the pen2 single mutant, the gsh1 single mutant showed a slight reduction in the preinvasive NHR against C. fioriniae CC1 and C. siamense MAF1 compared to other single mutantsCitation27 (). It was previously reported that GSH1 and PEN2 contribute to the same pathway in preinvasion NHR against HTE of C. tropicale S9275.Citation17 The slight effect of gsh1 single mutation against the MAE of these two fungi might partly reflect the link to the PEN2 pathway. GSH1 was of paramount importance for the resistance against fungal entry of C. fioriniae CC1, C. siamense MAF1, and C. siamense COC4 in the pen2 backgroundCitation27 (); however, in the case of C. nymphaeae PL1-1-b, the gsh1 single mutant did not show a higher MAE rate than the other single mutants, suggesting that the GSH1-related pathway is not very effective against C. nymphaeae PL1-1-b ().
Although PEN2, EDR1, GSH1, EDS5, CAS, and CHUP1 contribute to preinvasive NHR of Arabidopsis against Colletotrichum MAE, the preinvasive NHR of the edr1 pen2 gsh1 eds5 cas chup1 hexatruple mutant against C. orbiculare 104-T still has sufficient reservesCitation27 (). It is important to identify additional immune pathways that are effective against C. orbiculare 104-T and other untested nonadapted Colletotrichum strains that are highly incompatible with nonhost Arabidopsis.
PEN2 is a core NHR contributor to many fungal pathogens, including MAE-type P. oryzae and atypical HTE-type C. tropicale S9275.Citation5,Citation8,Citation13,Citation15,Citation21 Additionally, PEN2 conferred NHR against typical MAE-type Colletotrichum fungiCitation27 (). The use of nonadapted Colletotrichum fungi that can invade a single mutant, like pen2, through MAE, is promising; this will accelerate future research on NHR underlying Colletotrichum-Arabidopsis incompatible interactions. Currently, research on screening for NHR contributors against MAE of C. fioriniae CC1 is underway. In particular, the identification of immune component(s) other than PEN2, working in higher-layer preinvasive defenses is essential. C. nymphaeae PL1-1-b may avoid or suppress the function of such immune component(s) to partly overcome preinvasive NHR in the nonhost Arabidopsis.
Acknowledgments
I thank Paul Schulze-Lefert for pen2-1, Roger W. Innes for edr1-1, and Yoshitaka Takano for edr1-1 pen2-1, pen2-1 gsh1-1, edr1-1 pen2-1 gsh1-1, and Colletotrichum orbiculare (syn. C. lagenarium) 104-T (MAFF240422)
Disclosure statement
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Heath MC. Nonhost resistance and nonspecific plant defenses. Curr Opin Plant Biol. 2000;3(4):315–6. PMID: 10873843. doi:10.1016/s1369-5266(00)00087-x
- Lee HA, Lee HY, Seo E, Lee J, Kim SB, Oh S, Choi E, Choi E, Lee SE, Choi D. Current understandings of plant nonhost resistance. Mol Plant-Microbe Interact. 2017;30:5–15. PMID: 27925500. doi:10.1094/MPMI-10-16-0213-CR.
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–977. PMID: 14586469. doi:10.1038/nature02076.
- Jacobs AK, Lipka V, Burton RA, Panstruga R, Strizhov N, Schulze-Lefert P, Fincher GB. An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell. 2003;15(11):2503–2513. PMID: 14555698. doi:10.1105/tpc.016097
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310(5751):1180–1183. PMID: 16293760. doi:10.1126/science.1119409.
- Stein M, Dittgen J, Sánchez-Rodríguez C, Hou BH, Molina A, Schulze-Lefert P, Lipka V, Somerville S. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell. 2006;18:731–746. PMID: 16473969. doi:10.1105/tpc.105.038372.
- Jensen MK, Hagedorn PH, de Torres-zabala M, Grant MR, Rung JH, Collinge DB, Lyngkjaer MF. Transcriptional regulation by an NAC (NAM-ATAF1,2-CUC2) transcription factor attenuates ABA signalling for efficient basal defence towards Blumeria graminis f. sp. hordei in Arabidopsis. Plant J. 2008;56(6):867–880. PMID: 18694460. doi:10.1111/j.1365-313X.2008.03646.x
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science. 2009;323:101–106. PMID: 19095900. doi:10.1126/science.1163732.
- Pinosa F, Buhot N, Kwaaitaal M, Fahlberg P, Thordal-Christensen H, Ellerström M, Andersson MX. Arabidopsis phospholipase dδ is involved in basal defense and nonhost resistance to powdery mildew fungi. Plant Physiol. 2013;163(2):896–906. PMID: 23979971. doi:10.1104/pp.113.223503
- Campe R, Langenbach C, Leissing F, Popescu GV, Popescu SC, Goellner K, Beckers GJM, Conrath U. ABC transporter PEN3/PDR8/ABCG36 interacts with calmodulin that, like PEN3, is required for Arabidopsis nonhost resistance. New Phytol. 2016;209:294–306. PMID: 26315018. doi:10.1111/nph.13582.
- Nielsen ME, Jürgens G, Thordal-Christensen H. VPS9a activates the Rab5 GTPase ARA7 to confer distinct pre- and postinvasive plant innate immunity. Plant Cell. 2017;29(8):1927–1937. PMID: 28808134. doi:10.1105/tpc.16.00859
- Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, Humphry M, Bau S, Straus M, Kwaaitaal M, Rampelt H, et al. Co-option of a default secretory pathway for plant immune responses. Nature. 2008;451(7180):835–840. PMID: 18273019. doi:10.1038/nature06545.
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323(5910):95–101. PMID: 19095898. doi:10.1126/science.1164627
- Lu X, Dittgen J, Piślewska-Bednarek M, Molina A, Schneider B, Svatoš A, Doubský J, Schneeberger K, Weigel D, Bednarek P, et al. Mutant allele-specific uncoupling of PENETRATION3 functions reveals engagement of the ATP-binding cassette transporter in distinct tryptophan metabolic pathways. Plant Physiol. 2015;168(3):814–827. PMID: 26023163. doi:10.1104/pp.15.00182.
- Hiruma K, Onozawa-Komori M, Takahashi F, Asakura M, Bednarek P, Okuno T, Schulze-Lefert P, Takano Y. Entry mode-dependent function of an indole glucosinolate pathway in Arabidopsis for nonhost resistance against anthracnose pathogens. Plant Cell. 2010;22:2429–2443. PMID: 20605856. doi:10.1105/tpc.110.074344.
- Hiruma K, Nishiuchi T, Kato T, Bednarek P, Okuno T, Schulze-Lefert P, Takano Y. Arabidopsis ENHANCED DISEASE RESISTANCE 1 is required for pathogen-induced expression of plant defensins in nonhost resistance, and acts through interference of MYC2-mediated repressor function. Plant J. 2011;67(6):980–992. PMID: 21605210. doi:10.1111/j.1365-313X.2011.04651.x
- Hiruma K, Fukunaga S, Bednarek P, Pislewska-Bednarek M, Watanabe S, Narusaka Y, Shirasu K, Takano Y. Glutathione and tryptophan metabolism are required for Arabidopsis immunity during the hypersensitive response to hemibiotrophs. Proc Natl Acad Sci U S A. 2013;110(23):9589–9594. PMID: 23696664. doi:10.1073/pnas.1305745110
- Sels J, Mathys J, De Coninck BM, Cammue BP, De Bolle MF. Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol Biochem. 2008;46(11):941–950. PMID: 18674922. doi:10.1016/j.plaphy.2008.06.011
- Parisy V, Poinssot B, Owsianowski L, Buchala A, Glazebrook J, Mauch F. Identification of PAD2 as a gamma-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J. 2007;49:159–172. PMID: 17144898. doi:10.1111/j.1365-313X.2006.02938.x.
- Irieda H, Inoue Y, Mori M, Yamada K, Oshikawa Y, Saitoh H, Uemura A, Terauchi R, Kitakura S, Kosaka A, et al. Conserved fungal effector suppresses PAMP-triggered immunity by targeting plant immune kinases. Proc Natl Acad Sci U S A. 2019;116(2):496–505. PMID: 30584105. doi:10.1073/pnas.1807297116.
- Maeda K, Houjyou Y, Komatsu T, Hori H, Kodaira T, Ishikawa A. AGB1 and PMR5 contribute to PEN2-mediated preinvasion resistance to Magnaporthe oryzae in Arabidopsis thaliana. Mol Plant-Microbe Interact. 2009;22(11):1331–1340. PMID: 19810803. doi:10.1094/MPMI-22-11-1331
- Nakao M, Nakamura R, Kita K, Inukai R, Ishikawa A. Non-host resistance to penetration and hyphal growth of Magnaporthe oryzae in Arabidopsis. Sci Rep. 2011;1(1):171. PMID: 22355686. doi:10.1038/srep00171
- Okawa C, Ishikawa A. MPK6 contributes to non-host resistance to Magnaporthe oryzae in Arabidopsis thaliana. Biosci Biotechnol Biochem. 2013;77(6):1320–1322. PMID: 23748771. doi:10.1271/bbb.130082
- Takahashi T, Shibuya H, Ishikawa A. SOBIR1 contributes to non-host resistance to Magnaporthe oryzae in Arabidopsis. Biosci Biotechnol Biochem. 2016;80(8):1577–1579. PMID: 27023441. doi:10.1080/09168451.2016.1164586
- Takahashi T, Shibuya H, Ishikawa A. ERECTA contributes to non-host resistance to Magnaporthe oryzae in Arabidopsis. Biosci Biotechnol Biochem. 2016;80(7):1390–1392. PMID: 26924213. doi:10.1080/09168451.2016.1151345
- Takahashi T, Murano T, Ishikawa A. SOBIR1 and AGB1 independently contribute to nonhost resistance to Pyricularia oryzae (syn. Magnaporthe oryzae) in Arabidopsis thaliana. Biosci Biotechnol Biochem. 2018;82(11):1922–1930. PMID: 30022707. doi:10.1080/09168451.2018.1498727
- Irieda H, Takano Y. Epidermal chloroplasts are defense-related motile organelles equipped with plant immune components. Nat Commun. 2021;12:2739. PMID: 34016974. doi:10.1038/s41467-021-22977-5.
- Serrano M, Wang B, Aryal B, Garcion C, Abou-Mansour E, Heck S, Geisler M, Mauch F, Nawrath C, Métraux JP. Export of salicylic acid from the chloroplast requires the multidrug and toxin extrusion-like transporter EDS5. Plant Physiol. 2013;162:1815–1821. PMID: 23757404. doi:10.1104/pp.113.218156.
- Rekhter D, Lüdke D, Ding Y, Feussner K, Zienkiewicz K, Lipka V, Wiermer M, Zhang Y, Feussner I. Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science. 2019;365(6452):498–502. PMID: 31371615. doi:10.1126/science.aaw1720
- Nomura H, Komori T, Uemura S, Kanda Y, Shimotani K, Nakai K, Furuichi T, Takebayashi K, Sugimoto T, Sano S, et al. Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat Commun. 2012;3:926. PMID: 22735454. doi:10.1038/ncomms1926.
