ABSTRACT
Glutathione (GSH) is well known to play a crucial role in imparting resistance against various pathogen invasions. Nevertheless, the role of GSH in regulating miRNA-mediated defense response is yet to be explored. To decipher the GSH-mediated regulation of miRNA expression during necrotrophic infection in Arabidopsis thaliana, wild-type Col-0 and AtECS1, the transgenic line exhibiting enhanced GSH content, were infected with necrotrophic pathogen Alternaria brassicicola. AtECS1 plants exhibited enhanced resistance as compared to wild-type. MiRNA next-generation sequencing (NGS) was performed to compare the miRNA expression in Col-0 and AtECS1 leaves. Under control condition, differentially expressed 96 known miRNAs and 17 novel miRNAs viz. ath-miR8167f, ath-miR1886.3, ath-miR3932b-5p, etc. were identified. However, under infected condition, 73 known and 43 novel differentially expressed miRNAs viz. ath-miR5652, ath-miR160b, ath-miR865-5p, etc. were identified. Functional annotation and enrichment analysis revealed that several miRNAs that target defense-related genes like leucine-rich repeat protein kinase, MYB transcription factors, TCP8, etc. were down regulated in the AtECS1 line, which, in turn, relieves the repression of their target gene expression, leading to resistance against infection. Together, the present investigation suggests that GSH plays a decisive role in modulating the miRNA-mediated regulation of defense-related genes during pathogen invasion.
Introduction
In plant defense signaling network, glutathione (GSH) is gaining massive importance in disease resistance by playing a central role in regulating the defense signaling network in plants. In the earlier studies, it was shown that over-expression of GSH biosynthesis genes and increased GSH level can induce the expression of different disease-responsive genes, thus imparting stress tolerance.Citation1–3 Earlier, it was shown that GSH-deficient mutant pad2.1 was susceptible toward pathogen attack.Citation3 Exogenous application of GSH can also induce disease resistance in plants by activating NPR1-mediated SA signaling pathway, corroborating the regulatory role of GSH during biotic stress responses in plants.Citation1,Citation4
MicroRNAs (miRNAs) are a group of regulatory molecules with 21–24 nucleotides length and help in gene regulation by sequence-specific cleavage or translational repression of their target gene.Citation5 Several families of miRNAs have been identified from different plant species during pathogen attack, and their disease-responsive role has been investigated.Citation6–8 However, GSH-mediated regulation of any miRNAs during disease progression in plants is not studied so far. Alternaria brassicicola is a devastating necrotrophic fungal pathogen that causes dark spot disease in Brassicaceae family.Citation9 However, the role of GSH in regulating miRNA-mediated defense strategies in this pathosystem is still unexplored. In this study, our aim is to dissect the role of different miRNAs during A. brassicola infection in Arabidopsis and their regulation by GSH. Our study identified several stress-responsive miRNAs that are regulated by GSH during pathogen attack.
Materials and methods
Plant material and growth condition and pathogen inoculation
The transgenic A. thaliana line (AtECS1) overexpressing Le- γECS gene and exhibiting enhanced GSH content developed earlierCitation3 and Columbia-0 (Col-0) served as wild-type were used here. Plants were grown in Murashige and Skoog (MS) media and maintained in a growth chamber (22°C under 16 h light/8 h dark cycles).Citation10
Spore suspension of A. brassicicola strain MTCC No. 2102, obtained from MTCC Chandigarh, India, was used to inoculate leaves following van Wees et al..Citation11 For mock treatment, 5 μL droplets of water was used. Infection was scores at 5 dpi.
Trypan blue and DAB staining assay
To determine disease severity in Col-0 and AtECS1 plants, 4 weeks old Col-0 (CC), infected Col-0 (CI), AtECS1, and infected AtECS1 (AI) plants were used for trypan blue staining.Citation12 Infected leaves were put into 1:1 (vol/vol) ethanol and lactic acid–phenol–trypan blue solution for 45 min. Leaves were destained and mounted in 60% glycerol for observation.
DAB staining was performed according to Daudi et al.Citation13 Control and infected leaves were put into DAB solution (1 mg/ml) overnight and then the leaves were placed in bleaching solution (ethanol:acetic acid:glycerol = 3:1:1) for 5 min in boiling water bath and placed in fresh bleaching solution for 30 min and mounted for microscopic observation.
Small RNA library preparation and sequencing
Leaf from control and infected plants were used for the preparation of small RNA libraries. Total RNA was isolated using TRiZol reagent (Invitrogen, USA). The quality and quantity of RNA were analyzed using a Bioanalyser 2100 (Agilent Technologies), and a small RNA library was prepared using a small RNA Sample preparation Kit (Illumina Technologies, USA). The small RNA libraries were sequenced using a HiseqIllumina 1.5. The sequence data obtained as FASTQ files were assessed for qualitative analysis using FastQC version 3 and Fastx-toolkit, version 0.0.13.
Data mining and identification of miRNAs
The raw sequences generated were trimmed for removal of adaptor/primer contamination and poly(A) tails using an miRDeep adaptor filter. The pre-processed data were cleaned off including low quality reads. The unique reads (17–23 nucleotide) from two samples were retained for genome mapping. The filtered reads were screened against non-coding RNA sequences, tRNA, rRNA, and chloroplast sequences found in the Arabidopsis genome database. The reads homologous to these sequences were discarded. The miRDeep2 module was used to identify known and novel miRNAs in the high-throughput sequencing data.
Target gene prediction of novel miRNAs
EdgeR (R package, version 3.8.3) software was used to analyze the differential gene expression. The target gene was predicted using the psRNATarget server (http://plantgrn.noble.org/psRNATarget/) with default parameterCitation14,Citation15 and penalty score ≤2.5 .
Validation of miRNA expression and their corresponding target genes by qRT–PCR
Several putative novel and known miRNAs and their corresponding target genes were selected for qRT–PCR analysis. Isolation of miRNA was carried out using manufacturer’s protocol (Favorgen, Taiwan). The stem–loop and forward primers for the selected miRNAs were designed using miRNA primer designer software. Experiments were performed in three biological replicas.
Gene expression analysis of the predicted target genes were performed by qRT-PCR analysis in three biological replicas. The constitutively expressed elongation factor 1α gene was used as the reference gene. The primer pairs used is listed in Table S1.
Functional enrichment and annotation
Functional enrichment of differentially expressed miRNAs and their target genes was analyzed by singular enrichment analysis (SEA) with the agriGO tool.Citation16 SEA analysis computes GO term enrichment in the selected set of genes by comparing it to the reference set (in this case, TAIR genome locus TAIR10_2017). Fisher test and Benjamini-Yekutieli method was used here.
Results
Morphological analysis after A. brassicicola infection
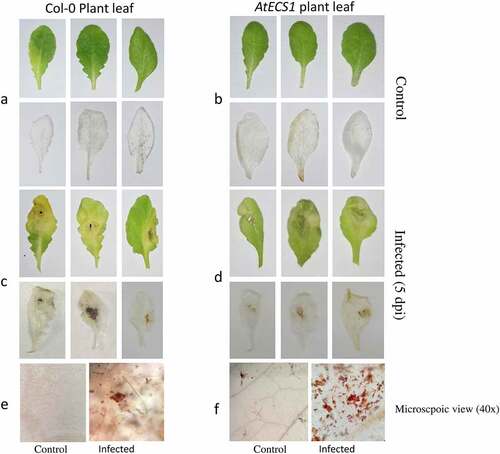
A. brassicicola-infected leaves showed dark necrotic spots. The Col-0 plants exhibited more susceptibility to infection compared to AtECS1 plants, suggesting that GSH plays a crucial role in imparting resistance in the AtECS1 plants against A. brassicicola infection.
Cell viability assessment and H2O2 detection after A. brassicicola infection
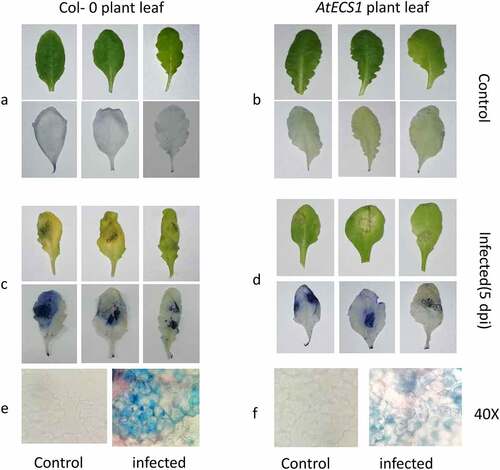
Trypan blue staining of Col-0 and AtECS1 plants infected with A. brassicicola revealed significantly more cell death in infected Col-0 than AtECS1 at 5 dpi () In DAB staining assay, the H2O2 accumulation in infected Col-0 leaves was also significantly higher than infected AtECS1, which once again indicates the role of GSH in making the plant more resistant against A. brassicicola infection ().
Figure 1. Trypan blue detection of necrotic cell in Col-0 and AtECS1 leaves after Alternaria brassicicola infection. a and b are controls (treated with ddH2O). No necrosis in leaf of Col-0 a and AtECS1 b at 5 dpi. c and d at 5 dpi with A. brassicicola. c Shows large necrotic area compare with d. e Microscopic view (40x) of control and infected leaf of Col-0. f microscopic view (40x) of control and infected leaf of AtECS1.
Identification of differentially expressed miRNA
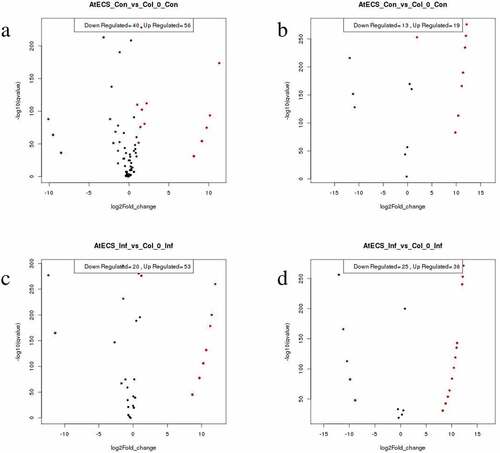
Sequencing of the CC, AC, CI, and AI leaf libraries generated a total of 507877, 391126, 179265, and 500809 sequence reads, respectively, provided in Table S2. After removal of low-quality sequence reads and adaptor contaminants, distinct sequences obtained were perfectly matched with the Arabidopsis genome. The differences in size compared with the total small RNA population were determined on the basis of total abundance reads. The read count of mature sequences of known and novel miRNAs of CC, AC, CI, and AI have been included in Table S3 and S4. MA-Plot diagram of the differential expression of miRNA in Col-0 and AtECS1 are given in . The mature sequences of novel miRNA found in all the 4 sets are included in Table S5. A total of 96 differentially expressed known and 17 novel miRNAs were identified in case of CC vs AC, and in case of CI vs AI, the total number of differentially expressed known miRNAs was 73 while the number of differentially expressed novel miRNAs identified was 43, which are provided in Table S6.
Figure 3. MA-plot diagram of the differential expression of miRNA in Col-0 and AtECS1. In red and green miRNAs differentially expressed with padj<0.05. a Known miRNA (AtECS1 Control vs Col-0 Control). b Novel miRNA (AtECS1 Control vs Col-0 Control). c Known miRNA (AtECS1 Infected vs Col-0 Infected). d Novel miRNA (AtECS1 Infected vs Col-0 Infected).
Validation of selected differentially expressed miRNAs and their target genes
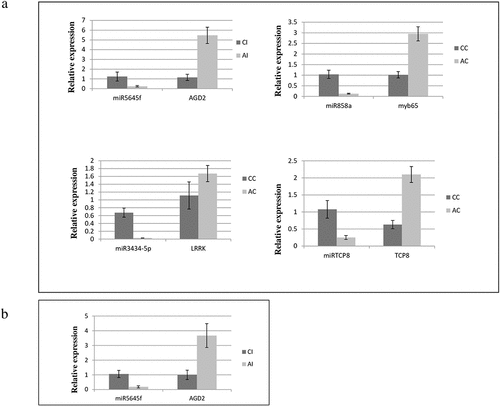
Few miRNAs like miR5645f, miR858a, miR3434-5p, and miRTCP8 expression data obtained from NGS analysis were confirmed through qRT-PCR analysis for several selected genes viz. AGD2, myb65, LRRK, and TCP8. The results indicated that the miRNAs were significantly differentially regulated in accordance with NGS results ().
Figure 4. Validation of differentially expressed known and novel miRNA and their respective target gene by qRT-PCR. U6 RNA gene was used as reference gene for miRNA and EF-1 alpha for target gene. Data are mean ± SD from three biological replicates determined by Student t test. a miRNA and their respective target under AtECS1 Control vs Col-0 Control (AC vs CC). b miRNA and their respective target under AtECS1 Infected vs Col-0 Infected (AI vs CI).
Functional enrichment and annotation
As observed by singular enrichment analysis by agriGo, the most significantly enhanced biological process for CC vs AC was regulation of transcription, followed by innate immune response, cellular response to stress, etc. In case of known miRNAs and in case of novel miRNAs, the most significantly enhanced biological process was defense response followed by response to stress (Table S7). In case of CI vs AI, most significant enhanced biological process was response to stimulus, metabolic process, transcription regulatory activity, immune system process, etc. (Table S8).
Discussion
A. brassicicola infection has detrimental effects on plants. However, plants response against this infection is yet to be explored in depth. Esa1, an Arabidopsis mutant with enhanced susceptibility to A. brassicicola, showed a distorted induction of defense responses by generating ROS.Citation17 Deficiency in phytoalexin production also showed enhanced susceptibility of planta to A. brassicicola.Citation18 Thus, previous studies on the A. brassicicola-Arabidopsis pathosystem can be explored further. Additionally, the enhanced level of GSH imparts significant tolerance to environmental stress,Citation19–24 and Nicotiana tabacum overexpressing γ-ECS exhibited biotic stress tolerance likely through NPR1-dependent salicylic acid-mediated pathway.Citation1 The present study showed that the A. thaliana overexpressing γ-ECS gene viz. AtECS1 showed increased resistance to A. brassicicola infection, depicting the important role of glutathione in conferring resistance to planta during biotic stress.
The discovery of miRNAs has led to a paradigm shift in understanding the post-transcriptional regulation of a gene in plants. Recent studies revealed that many miRNAs could be responsive to ethylene and play significant roles in plant growth and development.Citation25,Citation26 However, the direct role of miRNAs in controlling dynamic molecules like GSH is poorly understood. Present study focuses on the identification of miRNAs under altered GSH content in A. brassicicola-infected A. thaliana. To identify the GSH-responsive miRNAs under biotic stress, we constructed four miRNA leaf libraries from A. brassicicola-infected A. thaliana exhibiting altered GSH content. Deep sequencing of miRNAs revealed 96 known and 17 novel ones in CC and AC, whereas, 73 known and 43 novel miRNAs in CI and AI. In control samples, various known miRNAs playing diverse roles in biological processes like stress-responsive miRNAs like miR858a,Citation27 hormonal signaling pathways like miR5652, miR396a-3p etc.Citation15,Citation28 have been identified. In infected samples, various biotic stress responsive miRNAs like miR3932b-5p, mR5645f etc.Citation29 have also been identified. Most of the miRNAs were found to be downregulated in AC and AI with respect to CC and CI. Furthermore, 17 putative novel miRNAs were identified in CC vs AC. Out of this 17 novel miRNAs, 8 miRNAs were upregulated and 9 were downregulated. In CI vs AI samples, out of 43 putative novel miRNAs identified, 16 were downregulated and the rest were upregulated. Among the 96 known miRNAs in control samples of Arabidopsis, approximately 199 targets were predicted, and of the 17 novel miRNAs, almost 30 targets were predicted. Out of these predicted target genes, some were found to be functionally involved in defense response, and in case of infected A. thaliana having altered GSH level, the number of target gene found for known miRNAs were 307 and target genes for potential novel miRNAs were 68. These 68 target genes were found to be involved in multiple physiological and metabolism processes (Table S6). The expression profile from NGS data was validated by qRT-PCR. Few miRNAs and their target genes used are miR5645f-AGD2, miR858a-myb65, miR3434-5p-LRRK, and miRTCP8-TCP8. The result revealed that the miRNAs were significantly downregulated and their respective target genes were upregulated in AtECS1.
In order to better understand the potential role of miRNAs in A. thaliana, functional enrichment and annotation analysis was done by agriGO. Based on this analysis, many significant enriched GO terms were RNA biosynthetic process (GO:0032774) and transcriptional regulation (GO:0006355) in control samples of Arabidopsis. The significant GO terms in infected category were regulation of transcription (GO:0006355), regulation of response to biotic stimulus (GO:0009607), response to stress (GO:0006950), primary metabolic processes (GO:0044238), response to stimulus (GO:0050896), etc. Hence, in the presence of biotic stress, the miRNAs that were found to be downregulated in response to GSH induced the expression of its respective target genes, mainly LRRK, myb65, AGD2, and TCP8, which are involved in defense response and developmental processes (). Cumulatively, our findings suggest that under elevated GSH level in plants during pathogen attack, miRNAs regulate the stress-related genes and help the plant in combating stress.
Availability of data and material
The data described in this data note can be freely and openly accessed on the Sequence Read Archive (SRA) with Project ID GSE134863 in NCBI database and are also available from the corresponding author on reasonable request.
Authors’ contributions
RD and SC designed the experiments. RD carried out the major experimental work. PB and KM did further experiments and analyzed the data. AS help to analyse data. RD drafted the manuscript. SC supervised the analysis and prepared the final manuscript.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Supplemental Material
Download Zip (240.4 KB)Acknowledgments
The authors are grateful to the Director, CSIR-Indian Institute of Chemical Biology, Kolkata, for providing the necessary facilities. The authors would like to acknowledge CSIR and SERB, New Delhi, for financial support. Dr. SC would like to acknowledge CSIR, New Delhi, for Emeritus Scientist Fellowship. PB and AS and KM acknowledge the CSIR and SERB, New Delhi, for fellowships, respectively. The authors also acknowledge Nucleome Informatics Pvt. Ltd. (Hyderabad, India) for NGS data generation.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592324.2022.2047352
Disclosure statement
No potential conflict of interest was reported by the author(s).
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Ghanta S, Bhattacharyya D, Sinha R, Banerjee A, Chattopadhyay S. Nicotiana tabacum overexpressing γ-ECS exhibits biotic stress tolerance likely through NPR1-dependent salicylic acid-mediated pathway. Planta. 2011a;233(5):895–8. doi:10.1007/s00425-011-1349-4.
- Ghanta S, Bhattacharyya D, Chattopadhyay S. Glutathione signalling acts through NPR1-dependent SA-mediated pathway to mitigate biotic stress. Plant Signal Behav. 2011b;6(4):607–609. doi:10.4161/psb.6.4.15402.
- Datta R, Kumar D, Sultana A, Hazra S, Bhattacharyya D, Chattopadhyay S. Glutathione Regulates 1-Aminocyclopropane-1-Carboxylate Synthase Transcription via WRKY33 and 1-Aminocyclopropane-1-Carboxylate Oxidase by Modulating Messenger RNA Stability to Induce Ethylene Synthesis during Stress. Plant Physiol. 2015;169(4):2963–2981. doi:10.1104/pp.15.01543.
- Han Y, Chaouch S, Mhamdi A, Queval G, Zechmann B, Noctor G. Functional analysis of arabidopsis mutants points to novel roles for glutathione in coupling H 2 O 2 to activation of salicylic acid accumulation and Signaling. Antioxid Redox Signal. 2013;18(16):2106–2121. doi:10.1089/ars.2012.5052.
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5.
- Yadav A, Khan Y, Prasad M. Dehydration-responsive miRNAs in foxtail millet: genome-wide identification, characterization and expression profiling. Planta. 2016;243(3):749–766. doi:10.1007/s00425-015-2437-7.
- Zeng X, Xu Y, Jiang J, Zhang F, Ma L, Wu D, Wang Y, Sun W. Identification of cold stress responsive microRNAs in two winter turnip rape (Brassica rapa L.) by high throughput sequencing. BMC Plant Biol. 2018;18(1):52. doi:10.1186/s12870-018-1242-4.
- Wang TZ, Chen L, Zhao MG, Tian QY, Zhang WH. Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genomics. 2011;12(1):367. doi:10.1186/1471-2164-12-367.
- Lawrence CB, Mitchell TK, Craven KD, Cho YR, Cramer RA, Kim KH. At deaths door: alternaria pathogenicity mechanisms. Plant Pathol J. 2008;24(2):101–111. doi:10.5423/ppj.2008.24.2.101.
- Datta R, Sinha R, Chattopadhyay S. Changes in leaf proteome profile of Arabidopsis thaliana in response to salicylic acid. J Biosci. 2013;38(2):317–328. doi:10.1007/s12038-013-9308-9.
- van Wees Sc, Chang H-S, Zhu HS, Glazebrook J, van Wees SCM. Characterization of the early response of arabidopsis to alternaria brassicicola infection using expression profiling. Plant Physiol. 2003;132(2):606–617. doi:10.1104/pp.103.022186.
- Koch E, Slusarenko A. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell. 1990;2(5):437–445. doi:10.1105/tpc.2.5.437.
- Daudi A, O’Brien JA. Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio-protocol. 2012;2(18):e263. doi:10.21769/bioprotoc.263.
- Akpinar BA, Kantar M, Budak H. Root precursors of microRNAs in wild emmer and modern wheats show major differences in response to drought stress. Funct Integr Genomics. 2015;15(5):587–598. doi:10.1007/s10142-015-0453-0.
- Liu H, Searle IR, Watson-Haigh NS, Baumann U, Mather DE, Able AJ, Able JA. Genome-wide identification of microRNAs in leaves and the developing head of four durum genotypes during water deficit stress. PLoS One. 2015;10(11):e0142799. doi:10.1371/journal.pone.0142799.
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38(suppl_2):W64–W70.doi:10.1093/nar/gkq310. 30
- Tierens KF, Thomma BP, Bari RP, Garmier M, Eggermont K, Brouwer M, Penninckx IA, Broekaert WF, Cammue BP. Esa1, an Arabidopsis mutant with enhanced susceptibility to a range of necrotrophic fungal pathogens, shows a distorted induction of defense responses by reactive oxygen generating compounds. Plant J. 2002;29(2):131–140. doi:10.1046/j.1365-313x.2002.01199.x.
- Thomma BP, Nelissen I, Eggermont K, Broekaert WF. Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 1999;2(2):163–167. doi:10.1046/j.1365-313x.1999.00513.x.
- Noctor G, Arisi ACM, Jouanin L, Foyer CH. Manipulation of glutathione and amino acid biosynthesis in the chloroplast. Plant Physiol. 1998;118(2):471–482. doi:10.1104/pp.118.2.471.
- Gomez LD, Noctor G, Knight MR, Foyer CH. Regulation of calcium signaling and gene expression by glutathione. J Exp Bot. 2004;55(404):1851–1859. doi:10.1093/jxb/erh202.
- Gullner G, Komives T, Rennenberg H. Enhanced tolerance of transgenic poplar plants overexpressing γ-glutamylcysteine synthetase towards chloroacetanilide herbicides. J Exp Bot. 2001;52(358):971–979. doi:10.1093/jexbot/52.358.971.
- Xiang C, Werner BL, Christensen EM, Oliver DJ. The biological functions of glutathione revisited in Arabidopsis transgenic plants with altered glutathione levels. Plant Physiol. 2001;126(2):564–574. doi:10.1104/pp.126.2.564.
- Zhu YL, Pilon-Smits EAH, Tarun AS, Weber SU, Jouanin L, Terry N. Cadmium tolerance and accumulation in Indian mustard is enhanced by overexpressing c-glutamylcysteine synthetase. Plant Physiol. 1999;121(4):1169–1177. doi:10.1104/pp.121.4.1169.
- Herschbach C, van der Zalm E, Schneider A, Jouanin L, De Kok LJ, Rennenberg H. Regulation of sulfur nutrition in wild-type and transgenic poplar over-expressing γ-glutamylcysteine synthetase in the cytosol as affected by atmospheric H2S. Plant Physiol. 2000;124(1):461–473. doi:10.1104/pp.124.1.461.
- Chen L, Wang T, Zhao M, Zhang W. Ethylene-responsive miRNAs in roots of Medicago truncatula identified by high throughput sequencing at whole genome level. Plant Sci. 2011;184:14–19.doi:10.1016/j.plantsci.2011.11.007.
- Pei HX, Ma N, Chen JW, Zheng Y, Tian J, Li J, Zhang S, Fei ZJ, Gao JP, Bendahmane M. Integrative analysis of miRNA and mRNA profiles in response to ethylene in rose petals during flower opening. PLoS One. 2013;8(5):e64290. doi:10.1371/journal.pone.0064290.
- Camargo-ram�rez R, Val-Torregrosa B, San Segundo B. miR858-mediated regulation of flavonoid-specific MYB transcription factor genes controls resistance to pathogen infection in Arabidopsis. Plant Cell Physiol. 2018;59(1):190–204. 10.1371/journal.pone.0142799 et al., 2018] has been updated. OK?</chg>
- Curaba J, Singh MB, Bhalla PL. miRNAs in the crosstalk between phytohormone signalling pathways. J Exp Bot. 2014;65(6):1425–1438. doi:10.1093/jxb/eru002.
- Song JT, Lu H, Greenberg JT. Divergent roles in arabidopsis thaliana development and defense of two homologous genes, ABERRANT GROWTH AND DEATH2 and AGD2-LIKE DEFENSE RESPONSE PROTEIN1, Encoding Novel Aminotransferases. Plant Cell. 2004;16(2):353–366. doi:10.1104/pp.118.2.471.