ABSTRACT
WUSCHEL-RELATED HOMEOBOX 5 (WOX5) is a member of the WUSCHEL (WUS) homeodomain transcription factor family. WOX5 is expressed mainly in the quiescent center (QC) and confers stem cell identity in the root apical meristem (RAM). Consistent with the role of WUS in repressing root meristem development, we found that ectopic expression of WOX5 disrupted shoot development by repressing shoot-related genes, such as YABBY1 (YAB1). Our findings suggest that WOX5 and WUS potentially confer different tissue identities and specify RAM and SAM, respectively.
KEYWORDS:
Two primary meristems, shoot apical meristem (SAM) and root apical meristem (RAM), govern plant growth and development. Stem cell populations in the SAM and RAM are delicately modulated by a group of mitotically less-active cells comprising the organizing center (OC) and quiescent center (QC), respectively.Citation1,Citation2 Stem cell niches are established using a subset of WUSCHEL (WUS) family proteins. The WUS gene, which is specifically expressed in the OC, is responsible for SAM formation,1 whereas the QC-expressed WUSCHEL-RELATED HOMEOBOX 5 (WOX5) gene plays a key role in RAM formation.Citation3 Consistent with the origin of WUS and WOX5 proteins from a common ancestor,Citation4 both of these transcription factors perform a similar function in maintaining stem cell pluripotency, as shown by the complementation of wus mutants by pWUS::WOX5 expression or that of wox5 mutants by pWOX5::WUS.4
In addition to their overlapping functions, WUS and WOX5 have also been suggested to perform distinct functions, especially in tissue identity establishment. For example, conditional overexpression of the WUS gene leads to shoot organogenesis even from root tissues, possibly by generating shoot stem cell niche and also by inhibiting root meristem regulators such as PLETHORA 1 (PLT1).Citation5 These observations suggest the possibility that the WOX5 transcription factor is also suspected to affect shoot development, in addition to its original role in the maintenance of root stem cells.Citation6
To investigate the possibility that WOX5 could affect shoot development, we used transgenic Arabidopsis plants expressing the 35S::WOX5-GLUCOCORTICOID RECEPTOR (GR) construct,6 which enables dexamethasone (DEX)-induced nuclear targeting of the WOX5-GR fusion protein (). While wild-type seedlings were insensitive to DEX treatment and showed normal growth and development, regardless of DEX application, the DEX-treated 35S::WOX5-GR transgenic plants displayed dramatically disturbed shoot development (). In contrast, the ectopic expression of PLT2, which regulates root stem cell development similar to WOX5,Citation7 in 35S::PLT2-GR plants led to the production of normal shoots following DEX treatment unlike 35S::WOX5-GR, although overall shoot size was reduced (Supplemental Figure S1). These data suggest that WOX5 can suppress the shoot developmental program.
Figure 1. WOX5 inhibits shoot development. (a) Schematic representation of the study design. (b) Representative phenotype of dexamethasone (DEX)-treated 35S::WOX5-GR Arabidopsis seedlings. Wild-type and 35S::WOX5-GR plants were germinated on Murashige and Skoog (MS) medium supplemented with or without microM DEX, and grown under long-day (LD) conditions for 8 days. Scale bars = 1 mm. DAG, days after germination. (c) Percentage of shoot emergence. Cotyledon expansion was used as a phenotypic marker to evaluate shoot emergence. Statistically significant differences were determined using Student’s t-test (n= 33, ***P < 0.001). (d) Expression profiling of genes involved in SAM development. Total RNA was isolated from shoots of 11-day-old seedlings treated for 3 h with microM DEX or ethanol (EtOH). Transcript accumulation was analyzed by RT-qPCR. The eIF4a gene was used as an internal control. Data represent the mean ± standard error of the mean (SEM). Asterisks indicate statistically significant differences (***P < 0.001; Student’s t-test). (e) Schematic of WUS and WOX5 working model. OC-expressed WUS and QC-expressed WOX5 regulate the specification of SAM and RAM, respectively. In addition to their conserved function in pluripotency acquisition, WUS and WOX5 may also have potential roles in conferring distinct tissue identity. Ectopic activation of WUS inhibits root development genes even in roots and promotes shoot organogenesis from root tissues. In contrast, ectopic activation of WOX5 inhibits shoot development genes in shoots possibly through distinct interacting proteins that define tissue identity. SAM, shoot apical meristem; RAM, root apical meristem.
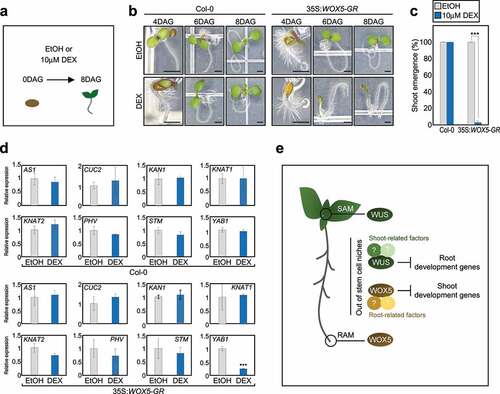
To gain insight into the role of WOX5 in shoot development, we analyzed expression of several genes involved in shoot development, including ASYMMETRIC LEAVES 1 (AS1), CUP-SHAPED COTYLEDON 2 (CUC2), KANADI 1 (KAN1), KNOTTED-LIKE FROM ARABIDOPSIS THALIANA 1 (KNAT1), KNAT2, PHAVOLUTA (PHV), SHOOT MERISTEMLESS (STM), and YABBY1 (YAB1),Citation8-10 in shoots of wild-type and 35S::WOX5-GR plants treated with DEX for 3 hours. Quantitative real-time RT-PCR (RT-qPCR) analysis revealed that DEX treatment specifically repressed YAB1 expression in 35S::WOX5-GR, but did not affect expression of other genes examined (). Given that YAB1 is a member of YABBY transcription factor family, which is specifically expressed in lateral organs of shoots,Citation11,Citation12 ectopic expression of WOX5 facilitated the inhibition of shoot developmental genes out of shoot stem cell niche. The regulation of YAB1 by WOX5 occurred possibly in SAM peripheral zone and abaxial domain of leaves, where YAB1 gene is mainly expressed.Citation11,Citation13,Citation14 We expect that WOX5 may further regulate a wide spectrum of shoot-expressed genes, in addition to YAB1, which should be examined in the future.
WOX5 and WUS, which originated from a common ancestor, exhibit conserved functions in the SAM and RAM, specifically during maintenance of stem cell niches in these meristematic tissues.4 Complementation of wus-1 by WOX5 expression driven by the native WUS promoter corrects the defect in SAM development.4 Similarly, pWOX5::WUS expression rescues abnormal RAM development in wox5 mutants.4 While the overlapping functions of WUS and WOX5 are reflected in stem cell niches, they also potentially have unique functions. Ectopic expression of WUS inhibits root development possibly via suppression of root stem cell regulators.5 Our results also showed that ectopic expression of WOX5 inhibits shoot development probably by repressing shoot-related genes. Since WUS and WOX5 complement functions each other in stem cell niches,4 their opposite functions in tissue identity establishment are most likely facilitated in the regions outside of stem cell niches, which might be related to the maintenance of body axis. Outside of stem cell niches, WUS and WOX5 may have more chance to interact with shoot-related and root-related proteins, respectively, which repress opposite tissue identity. Indeed, the unique protein interactome of each protein was suspected,Citation15 and a list of interacting proteins of WOX5 and WUS were related to tissue-specific factors (Supplemental Figure S2), suggesting that spatial mis-expansion may lead to new regulatory repertoires via distinct molecular interactions.Citation16 We could raise the possibility that the spatial mis-expression caused biological artifacts. Nonetheless, the genetic impact needs to be understood and can sometimes also be applied to artificial control of plant development. Overall, we propose that, while WUS and WOX5 exhibit a conserved function owing to their common ancestor, their subsequent functional divergence might have led to evolution of their tissue-specific roles in the SAM and RAM, respectively.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia (Col-0) was used in all experiments, unless specified otherwise. Plants were grown at 22–23°C under long-day (LD) conditions (16 h light/8 h dark) using white fluorescent lamps (150 μmol photons m−2s−1). For DEX treatment, 35S::WOX5-GR6 and 35S::PLT2-GRCitation17 seeds were germinated in Murashige and Skoog (MS) medium supplemented with or without 10 μM DEX.
RT-qPCR analysis
Total RNA was extracted from the plant materials of interest using the TRI Reagent (Takara Bio), according to the manufacturer’s instructions. First-strand cDNA was synthesized from 2 μg of total RNA using Moloney Murine Leukemia Virus reverse transcriptase (Dr. Protein) and dT20 oligos. The cDNA was diluted to a volume of 100 μl with Tris-EDTA (TE) buffer, and 1 μl of the diluted cDNA was used for RT-qPCR.
The RT-qPCR reactions were performed on 96-well plates using the StepOnePlus Real-Time PCR System (Applied Biosystems). Gene expression levels were normalized relative to that of EUKARYOTIC TRANSLATION INITIATION FACTOR 4A1 (eIF4A; At3g13920). The primers used for RT-qPCR are listed in Supplemental Table S1. The relative gene expression levels were quantified using the comparative ΔΔCt method. The threshold cycle (Ct) for each reaction was determined automatically by the analysis software using default parameters (Applied Biosystems). The specificity of RT-qPCR reactions was determined by melting curve analysis.
Supplemental Material
Download PDF (75.6 KB)Acknowledgments
We thank Dr. Renze Heidstraa (Wageningen University and Research Centre, Netherlands) and Dr. Thomas Laux (Albert-Ludwigs-University Freiburg, Germany) for kindly providing the 35S::PLT2-GR and 35S::WOX5-GR transgenic seeds, respectively.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
Additional information
Funding
References
- Tucker MR, Laux T. Connecting the paths in plant stem cell regulation. Trends Cell Biol. 2007;17:403–3. doi:10.1016/j.tcb.2007.06.002.
- Gaillochet C, Lohmann JU. The never-ending story: from pluripotency to plant developmental plasticity. Development. 2015;142:2237–2249. doi:10.1242/dev.117614.
- Kong X, Lu S, Tian H, Ding Z. WOX5 is shining in the root stem cell niche. Trends Plant Sci. 2015;20:601–603. doi:10.1016/j.tplants.2015.08.009.
- Zhang Y, Jiao Y, Jiao H, Zhao H, Zhu YX. Two-step functional innovation of the stem-cell factors WUS/WOX5 during plant evolution. Mol Biol Evol. 2017;34:640–653. doi:10.1093/molbev/msw263.
- Negin B, Shemer O, Sorek Y, Eshed Williams L. Shoot stem cell specification in roots by the WUSCHEL transcription factor. PLoS One. 2017;12:e0176093. doi:10.1371/journal.pone.0176093.
- Pi L, Aichinger E, van der Graaff E, Llavata-Peris CI, Weijers D, Hennig L, Groot, E, Laux, T . Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev Cell. 2015;33:576–588. doi:10.1016/j.devcel.2015.04.024.
- Ding Z, Friml J. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc Natl Acad Sci USA. 2010;107:12046–12051. doi:10.1073/pnas.1000672107.
- Sarojam R, Sappl PG, Goldshmidt A, Efroni I, Floyd SK, Eshed Y, Bowman, JL . Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell. 2010;22:2113–2130. doi:10.1105/tpc.110.075853.
- Kim SY, Lee J, Eshed-Williams L, Zilberman D, Sung ZR. EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genet. 2012;8:e1002512. doi:10.1371/journal.pgen.1002512.
- Tsukaya H. Leaf development. Arabidopsis Book. 2013;11:e0163. doi:10.1199/tab.0163.
- Goldshmidt A, Alvarez JP, Bowman JL, Eshed Y. Signals derived from YABBY gene activities in organ primordia regulate growth and partitioning of Arabidopsis shoot apical meristems. Plant Cell. 2008;20:1217–1230. doi:10.1105/tpc.107.057877.
- Zhang S, Wang L, Sun X, Li Y, Yao J, van Nocker S, Wang X. Genome-wide analysis of the YABBY gene family in Grapevine and functional characterization of VvYABBY4. Front Plant Sci. 2019;10:1207. doi:10.3389/fpls.2019.01207.
- Nakata M, Matsumoto N, Tsugeki R, Rikirsch E, Laux T, Okada K. Roles of the middle domain-specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell. 2012;24:519–535. doi:10.1105/tpc.111.092858.
- Bonaccorso O, Lee JE, Puah L, Scutt CP, Golz JF. FILAMENTOUS FLOWER controls lateral organ development by acting as both an activator and a repressor. BMC Plant Biol. 2012;12:176. doi:10.1186/1471-2229-12-176.
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D13. doi:10.1093/nar/gky1131.
- Romanova MA, Maksimova AI, Pawlowski K, Voitsekhovskaja OV. YABBY genes in the development and evolution of land plants. Int J Mol Sci. 2021;23:22. doi:10.3390/ijms23010022.
- Santuari L, Sanchez-Perez GF, Luijten M, Rutjens B, Terpstra I, Berke L, Gorte M, Prasad K, Bao D, Timmermans-Hereijgers JLPM, et al. The PLETHORA gene regulatory network guides growth and cell differentiation in Arabidopsis roots. Plant Cell. 2016;28:2937–2951. doi:10.1105/tpc.16.00656.
